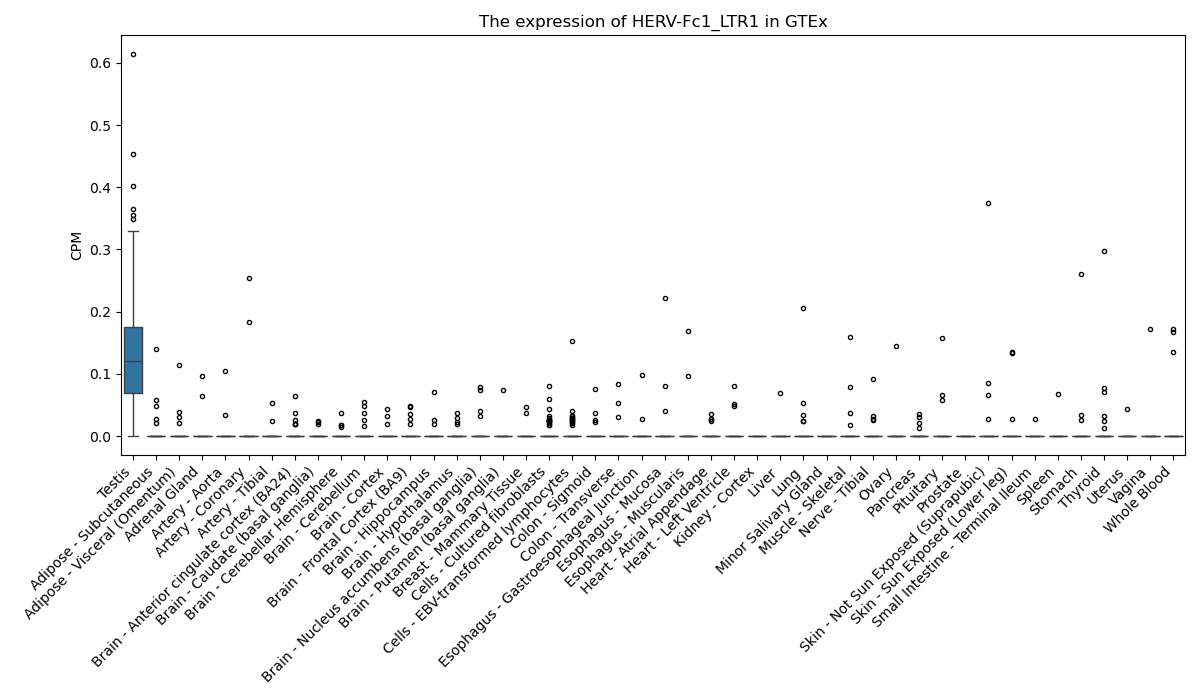
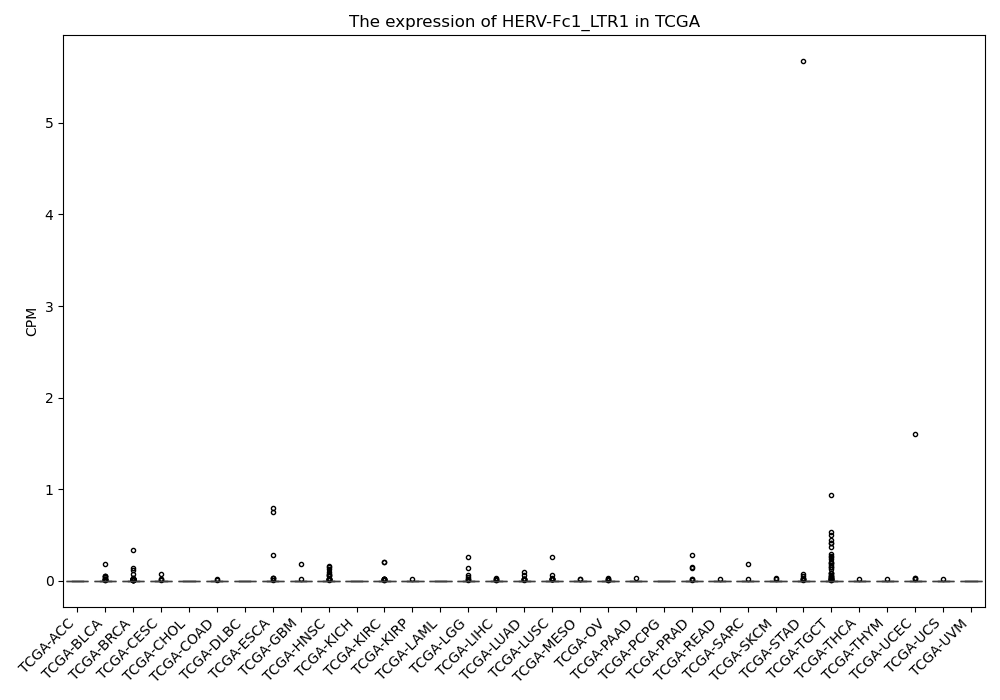
HERV-Fc1_LTR1
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000177 |
|---|---|
| TE superfamily | ERV1 |
| TE class | LTR |
| Species | Homininae |
| Length | 346 |
| Kimura value | 2.12 |
| Tau index | 1.0000 |
| Description | LTR1 (Long Terminal Repeat) for endgenous retrovirus HERV-Fc1 |
| Comment | - |
| Sequence |
TGATTGGACTCCGGTGAAGACTCCAAGATGGCGATCGCCACCTCGGATACCCTGACTCAGCATTTCCGGGTTCACCTTTCCTGTTCCCGCCACCCCGACTAACGCGCATGCCCACTAGGGCGTGTCACACTCAGAAGCGCGAAACTCAACCGACCCCGCCCCTACCCCGCCCACTCCTCACCCAGCATCCATAAAAGCGCGCTGCACCTTTGGCACAGCGCGACTTCCCTGGCCCTCCCCCTGCGGACCAGTGAACCTCGCCCGAGAGCTCAATAAAGAAGATTTTTGCCCTCTTTGTCTCGCCTCTCGGCCTTATTGATCCACGGTGCCCTTCCATTGCCTTTCA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| HERV-Fc1_LTR1 | KLF1 | 154 | 161 | - | 15.53 | GGGCGGGG |
| HERV-Fc1_LTR1 | KLF1 | 165 | 172 | - | 15.53 | GGGCGGGG |
| HERV-Fc1_LTR1 | SP4 | 165 | 173 | - | 15.52 | TGGGCGGGG |
| HERV-Fc1_LTR1 | ELF2 | 62 | 71 | - | 15.34 | ACCCGGAAAT |
| HERV-Fc1_LTR1 | KLF11 | 165 | 174 | + | 15.33 | CCCCGCCCAC |
| HERV-Fc1_LTR1 | MAFG::NFE2L1 | 52 | 62 | + | 15.30 | CTGACTCAGCA |
| HERV-Fc1_LTR1 | E2F6 | 84 | 91 | - | 15.29 | GGCGGGAA |
| HERV-Fc1_LTR1 | Spps | 153 | 163 | + | 15.20 | ACCCCGCCCCT |
| HERV-Fc1_LTR1 | Sp1 | 166 | 174 | + | 15.00 | CCCGCCCAC |
| HERV-Fc1_LTR1 | KLF16 | 164 | 174 | + | 14.98 | ACCCCGCCCAC |
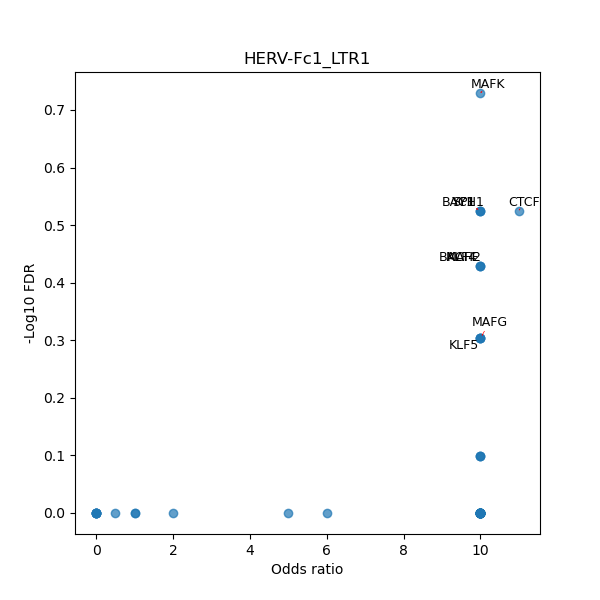
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.