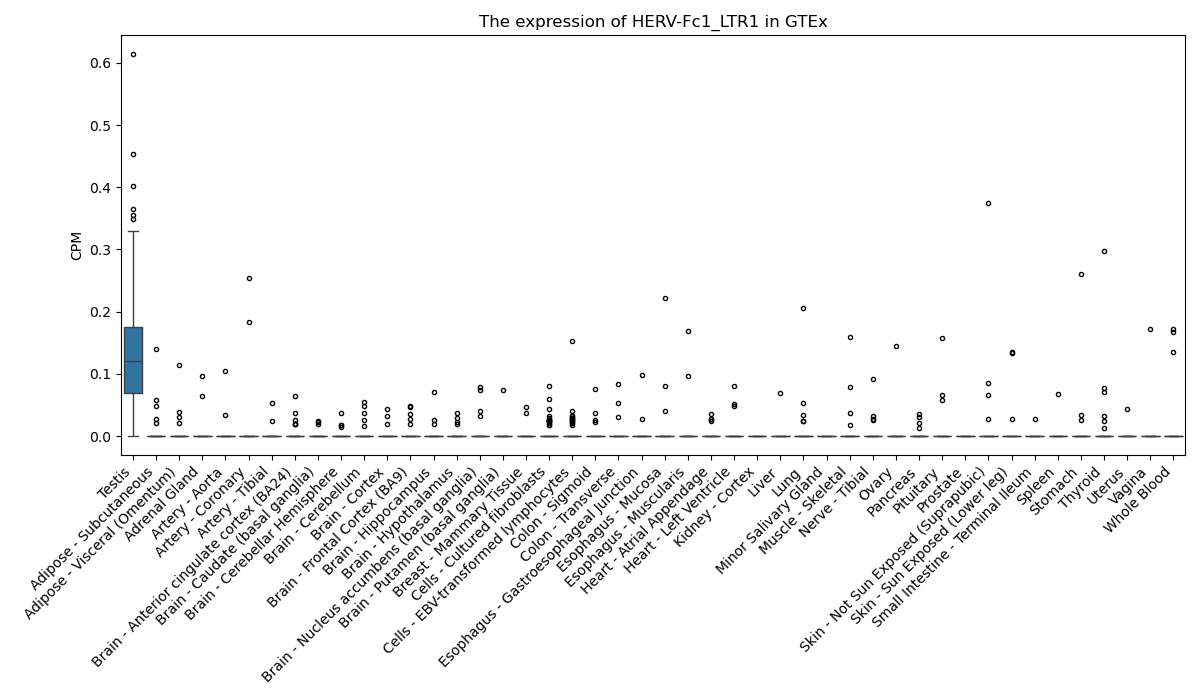
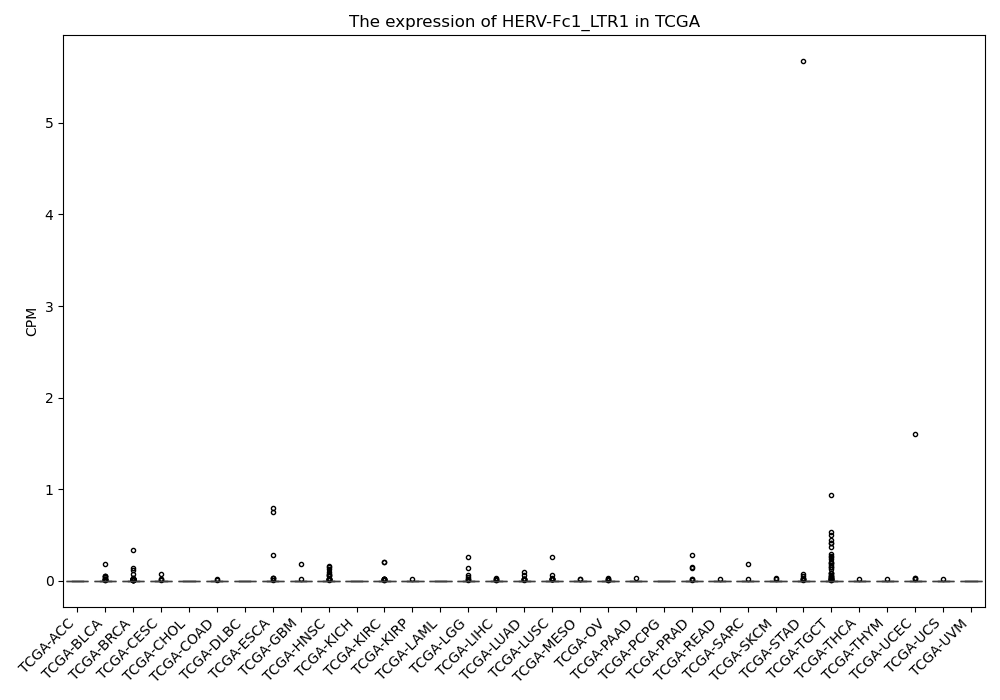
HERV-Fc1_LTR1
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000177 |
|---|---|
| TE superfamily | ERV1 |
| TE class | LTR |
| Species | Homininae |
| Length | 346 |
| Kimura value | 2.12 |
| Tau index | 1.0000 |
| Description | LTR1 (Long Terminal Repeat) for endgenous retrovirus HERV-Fc1 |
| Comment | - |
| Sequence |
TGATTGGACTCCGGTGAAGACTCCAAGATGGCGATCGCCACCTCGGATACCCTGACTCAGCATTTCCGGGTTCACCTTTCCTGTTCCCGCCACCCCGACTAACGCGCATGCCCACTAGGGCGTGTCACACTCAGAAGCGCGAAACTCAACCGACCCCGCCCCTACCCCGCCCACTCCTCACCCAGCATCCATAAAAGCGCGCTGCACCTTTGGCACAGCGCGACTTCCCTGGCCCTCCCCCTGCGGACCAGTGAACCTCGCCCGAGAGCTCAATAAAGAAGATTTTTGCCCTCTTTGTCTCGCCTCTCGGCCTTATTGATCCACGGTGCCCTTCCATTGCCTTTCA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| HERV-Fc1_LTR1 | THRB | 317 | 334 | + | 3.50 | TGATCCACGGTGCCCTTC |
| HERV-Fc1_LTR1 | THRB | 72 | 89 | + | 3.36 | TCACCTTTCCTGTTCCCG |
| HERV-Fc1_LTR1 | ZNF16 | 287 | 307 | - | 2.85 | AGAGGCGAGACAAAGAGGGCA |
| HERV-Fc1_LTR1 | E2FD | 82 | 96 | - | -0.16 | GGGGTGGCGGGAACA |
| HERV-Fc1_LTR1 | E2FE | 82 | 96 | - | -0.16 | GGGGTGGCGGGAACA |
| HERV-Fc1_LTR1 | E2F7 | 82 | 95 | + | -5.18 | TGTTCCCGCCACCC |
| HERV-Fc1_LTR1 | Rarg | 317 | 333 | - | -6.31 | AAGGGCACCGTGGATCA |
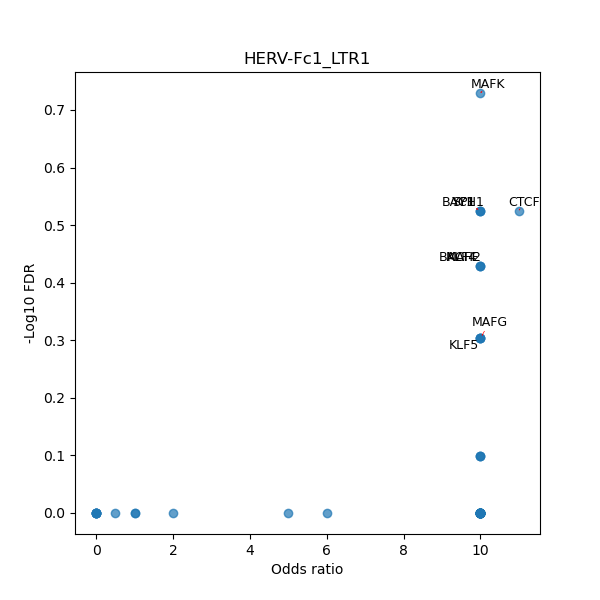
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.