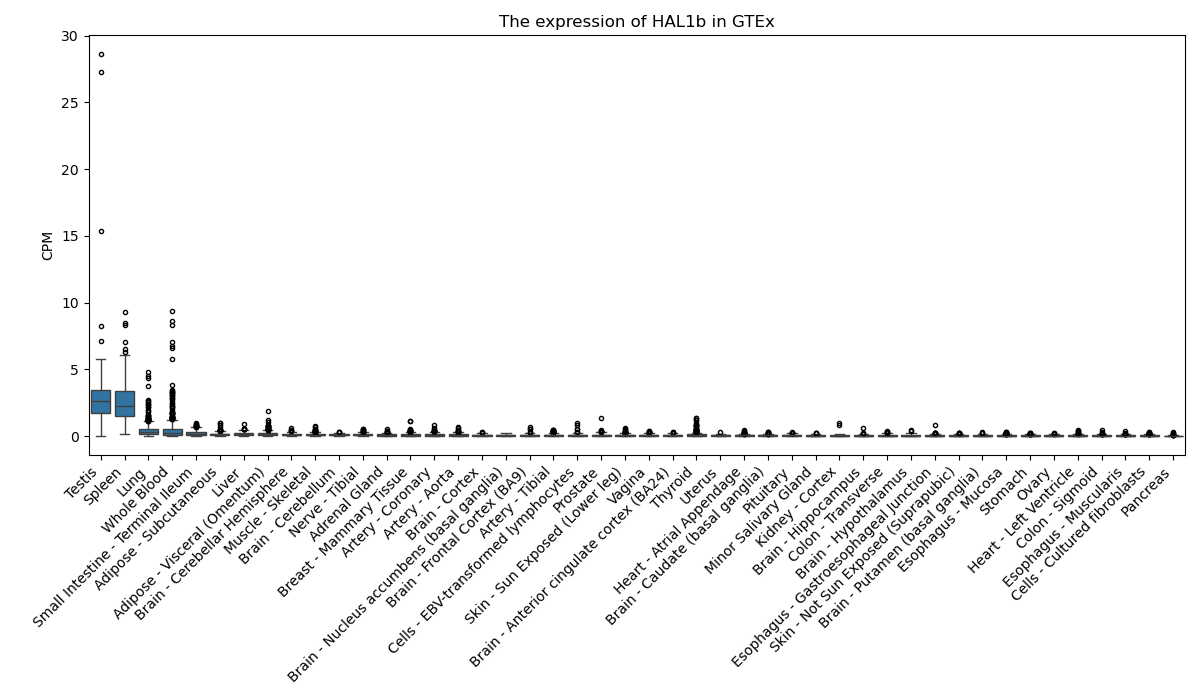
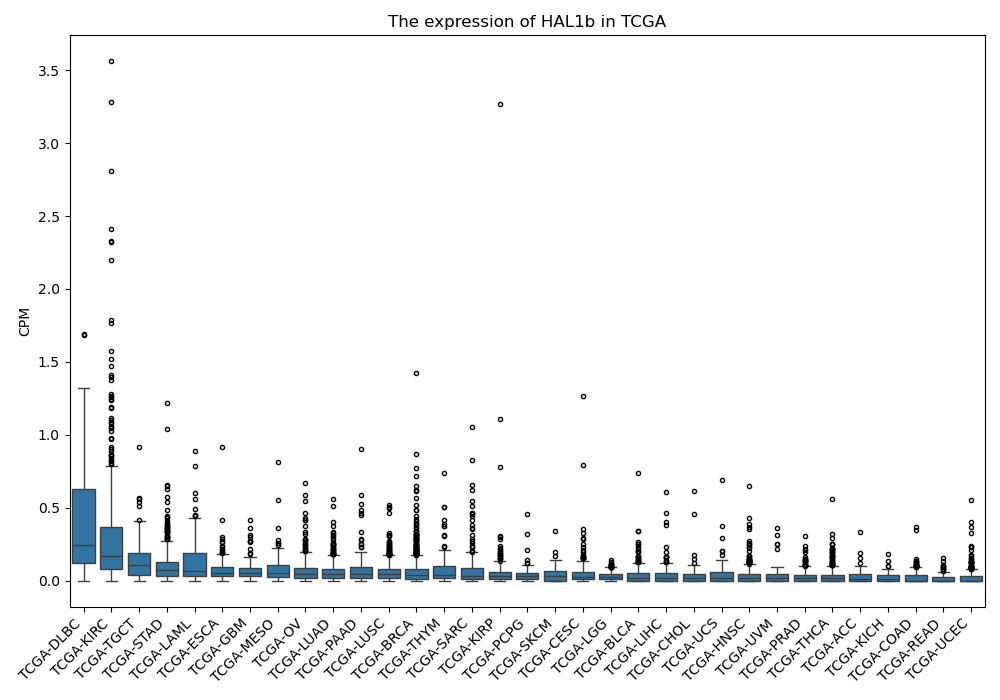
HAL1b
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000155 |
|---|---|
| TE superfamily | L1 |
| TE class | LINE |
| Species | Eutheria |
| Length | 2421 |
| Kimura value | 30.63 |
| Tau index | 0.9553 |
| Description | L1 retrotransposon, HAL1b subfamily |
| Comment | The 'ORF1-like' coding region ends somewhere around bp 660. gag ORF from 715-1686. |
| Sequence |
AAAAATTCACCAGCAGCAACCAAACAAGGAAAGGGGTACAACCTCATGCCATAAACTTCGAAAAGTGGCGGCCAAGGAAAGAAACAGAAGATTCTGGCGCGGTNTAGCATGGCTCCTAACCCCGCCCCCACCCCCCAGAAGCGGTACTGAGGAAAGAAGGTGAGTCGACAAAATCCTGAGAATTCTCACTAATACCCTCCAATTTGGAGAGAAGCGGACTGAGGGGGTGAGTAGGCAGCCCNGGGAAAAGTCAAGGAAAAACCCTCGAGAGTAGAAGCCCCTACGTACCACCGCGGGACCTCGGCAGAGGCAGGAAGGGCCGCCAGGGCTTGCCATCTTCCAGGGCNCCGCGCTGAATTGGGGGAATGANCCGAAGCCCACGGGAGTGGGATACGCCCCCGGCTAGGTGGAGTGAGCCCTGGTACAGGACATTTAACGAAATCTCAGAAGAGAGAGCAGAGCCCAGGTCCTCCAACAAAGGCTGCACNCAGCCCCAGCCGAGCTCTCCCCCCTTCCCCCATCCCCGCGCCCTCAGGCTACAGAAAAGTGGATAGAAAGCAATACCCAGCCCTCAAACTGTAGCTCAGAGGGGAAGACAAACAGGTCTCAGATAAGGTGGGAAGGGNGTCAAGGAGAATTCACCCACACTATATCAGAGCAAATAGAAGGTCTGGACAGAGCTGCCCACGTTAAGCATGAGCAAGATGAAAAGAAATGGCATGGCAACTATGCAAACATACTACAGCAAAGAAGAGAGAGNGAAAGGAAGAAAGGAGACANAGCATGCAAAAGACAGACGAAGAACCCAGTCTGGAAGAAAACATAACCCAAGGAACAGAAGAAAATTCCCCGCGAGTGCTTCGCGCTATAGAANAACTTAATAAGAACATGAATTCAATAAAACAAGAGCTCAAAGACGAGATGATAAAACAACAGAATGAGATGAAAAGGGAGATTGCAGAGCTAAGGAAACAAATTGAGGACCAAAACAACACCATTACAGAACTAATAAATAAATTAGAAACAGCAAGGAACAGAATAGACACGGCTGAAAATCGAATTACTGACATAGAGGAAAGGCTTGAGATAATCACAGTGAATGCAGAAGGAAAAGACAAAGAGATTAAAGCAATTAGAGAGAAGATAATAGATATGGAGGACAGACAAAGACGATCCAACATAAGGATAATTGGTGTCCCTGAAGTAGAGAACCCAACAAATGGAACAGAAAAAGTATTCAAAGATATAATACAAGAAAATTTCCCTGAAATGAAGGAAGAACTGAATCTGCAGATCGAAAGGGCACACCGTGTTCCAGGAAAAATTGATACAGAACGATCGACACCGAGACATATCCTGGTTAAGTTATTGAACTTCAAGGATAAAGAAAGAATTCTTCAGGCATCCAGGCAGAAAAAGCAAGTCACCTACAAGGGGGAAAAAATCAGGCTGGCCTCAGACTTCTCCACAGCAACATTCAATGCCAGAAGACAATGGAGCAATGTCTACAAAGTTCTGAGGGAAAGAAAGTGTGACCCAAGAATNTTATACCCAGCCAAGTTGTCGTTCAAGTATAAAGGCAACAGGCAGACATTCTCAAACATGAAAGAACTCAGGGAATATAGCACCCATGAGCCCTTCTTGAAAAAACTACTTGACGATGAAATCCAGCCAACCAAGAGATGAATCAAAATAAAGAACTCAGGAATGGAGAAGCCGTGGTAAAAGGACTGGTGGTGAGCACTGAATCCATTTAAATATAGAACTAAGACTAAACAACTGTGGGAATTATGGTTACAGAACAGAATGTAAATGTTATAAACCTTGACAANGTAAAAATAATATAANTAACAAAAATCGGGAGGTGGGAGAGGAGAGGTGGGAGGAAGTGTAAGAGTGCTAATTTCCTCATCTTTCATAGCAGGGAGTCAATCGATACTGTCTAAAATTGAAACATGTAGTTTAAAAANATAATGACTCCAACCTCTTAATGTTTTTCATAATCTTTTTTCTTAACCTTAGAGGGATCTTTTAGGAANTAATATCTCTTGTGGTGAAGAAACATTTATCTGAAGTTCAGCAATTCCTTCAGTTTCACTTCAGTTTCTTTTTCTTCTGTTAAATTCAAGTAAAATTAAATTTAATACTTTTNATTTAAAATAGCATGTATAGTATGATCCCATTTTTATAAAATTATTTCTATCTATCCATCCATCCATCCATCTATCCTTCTATCTGTATATATGCATAGAAAGATGTCTGGAATGATGTTCACCAAATGTTAACGATGGTTATTTCTGGGTGGTGGGATTTNGGGTGATTTNTTGCTTTCTTCTTTGTACTTTTCTGTATTGCTTGAATTTTTTATAATGAGCATGTATCATTTTTATAAAAACAATAAAGTNATTATTTTTTTAAAAA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| HAL1b | Spps | 119 | 129 | + | 16.97 | ACCCCGCCCCC |
| HAL1b | PRDM9 | 116 | 135 | - | 16.88 | GGGGGTGGGGGCGGGGTTAG |
| HAL1b | SP3 | 119 | 129 | + | 16.85 | ACCCCGCCCCC |
| HAL1b | KLF16 | 119 | 129 | + | 16.69 | ACCCCGCCCCC |
| HAL1b | DOF5.8 | 2329 | 2347 | + | 16.64 | TTTCTTCTTTGTACTTTTC |
| HAL1b | WRKY50 | 246 | 254 | - | 16.45 | TTGACTTTT |
| HAL1b | BAD1 | 120 | 131 | + | 16.34 | CCCCGCCCCCAC |
| HAL1b | SP9 | 120 | 129 | + | 16.33 | CCCCGCCCCC |
| HAL1b | KLF15 | 120 | 127 | + | 16.27 | CCCCGCCC |
| HAL1b | Klf15 | 116 | 137 | + | 16.14 | CTAACCCCGCCCCCACCCCCCA |
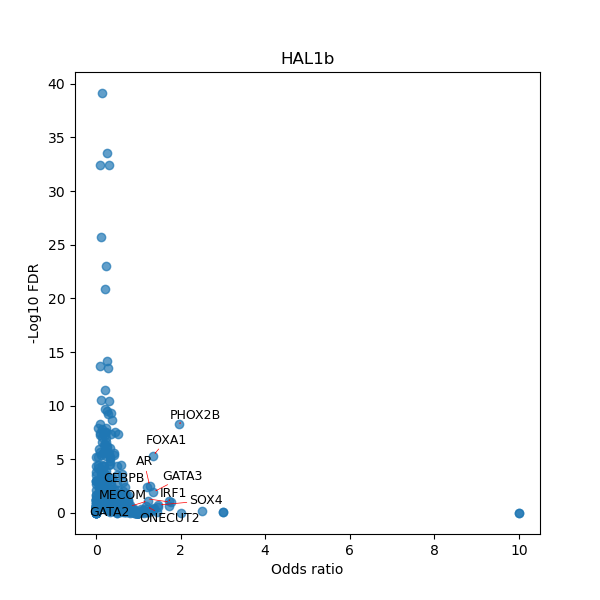
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.