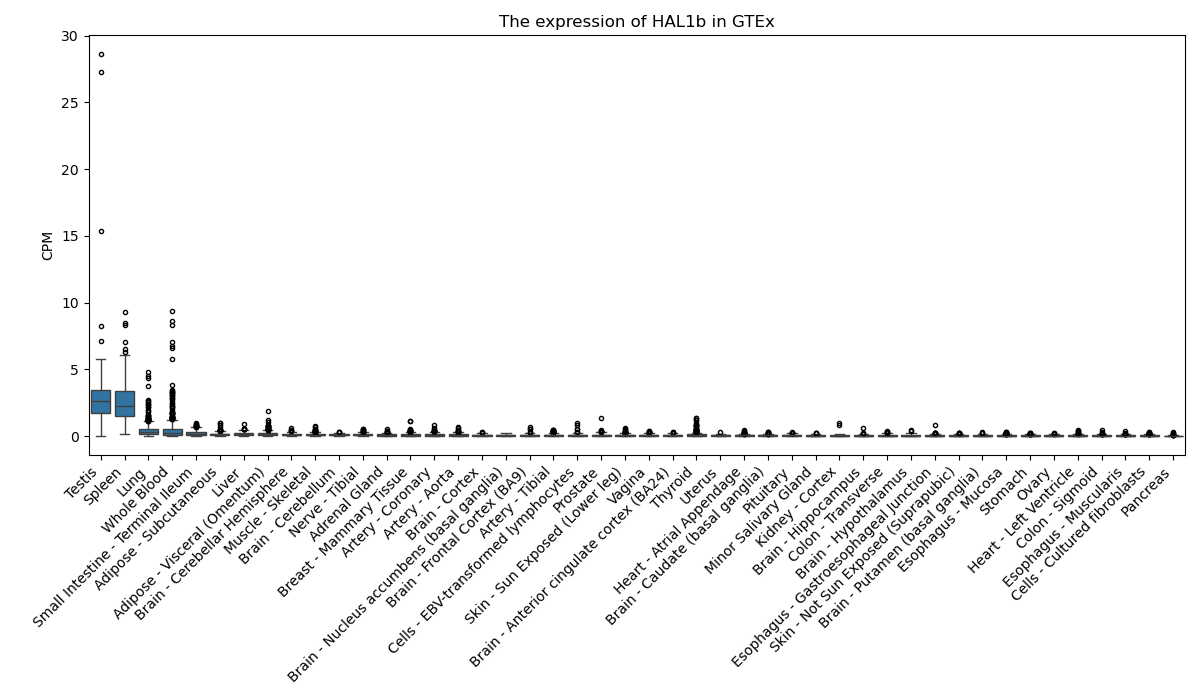
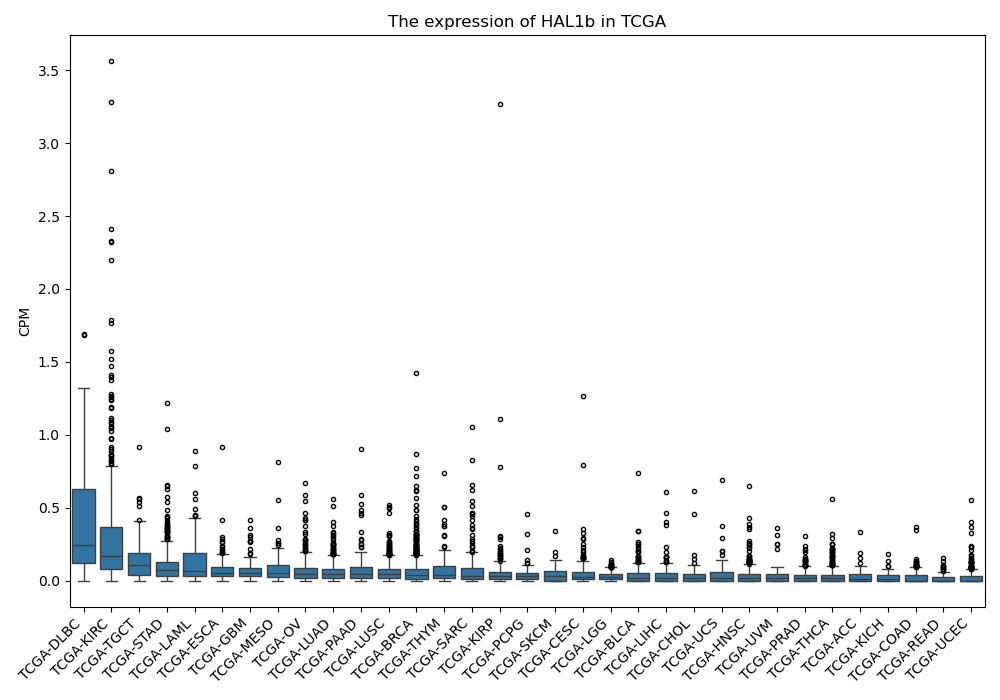
HAL1b
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000155 |
|---|---|
| TE superfamily | L1 |
| TE class | LINE |
| Species | Eutheria |
| Length | 2421 |
| Kimura value | 30.63 |
| Tau index | 0.9553 |
| Description | L1 retrotransposon, HAL1b subfamily |
| Comment | The 'ORF1-like' coding region ends somewhere around bp 660. gag ORF from 715-1686. |
| Sequence |
AAAAATTCACCAGCAGCAACCAAACAAGGAAAGGGGTACAACCTCATGCCATAAACTTCGAAAAGTGGCGGCCAAGGAAAGAAACAGAAGATTCTGGCGCGGTNTAGCATGGCTCCTAACCCCGCCCCCACCCCCCAGAAGCGGTACTGAGGAAAGAAGGTGAGTCGACAAAATCCTGAGAATTCTCACTAATACCCTCCAATTTGGAGAGAAGCGGACTGAGGGGGTGAGTAGGCAGCCCNGGGAAAAGTCAAGGAAAAACCCTCGAGAGTAGAAGCCCCTACGTACCACCGCGGGACCTCGGCAGAGGCAGGAAGGGCCGCCAGGGCTTGCCATCTTCCAGGGCNCCGCGCTGAATTGGGGGAATGANCCGAAGCCCACGGGAGTGGGATACGCCCCCGGCTAGGTGGAGTGAGCCCTGGTACAGGACATTTAACGAAATCTCAGAAGAGAGAGCAGAGCCCAGGTCCTCCAACAAAGGCTGCACNCAGCCCCAGCCGAGCTCTCCCCCCTTCCCCCATCCCCGCGCCCTCAGGCTACAGAAAAGTGGATAGAAAGCAATACCCAGCCCTCAAACTGTAGCTCAGAGGGGAAGACAAACAGGTCTCAGATAAGGTGGGAAGGGNGTCAAGGAGAATTCACCCACACTATATCAGAGCAAATAGAAGGTCTGGACAGAGCTGCCCACGTTAAGCATGAGCAAGATGAAAAGAAATGGCATGGCAACTATGCAAACATACTACAGCAAAGAAGAGAGAGNGAAAGGAAGAAAGGAGACANAGCATGCAAAAGACAGACGAAGAACCCAGTCTGGAAGAAAACATAACCCAAGGAACAGAAGAAAATTCCCCGCGAGTGCTTCGCGCTATAGAANAACTTAATAAGAACATGAATTCAATAAAACAAGAGCTCAAAGACGAGATGATAAAACAACAGAATGAGATGAAAAGGGAGATTGCAGAGCTAAGGAAACAAATTGAGGACCAAAACAACACCATTACAGAACTAATAAATAAATTAGAAACAGCAAGGAACAGAATAGACACGGCTGAAAATCGAATTACTGACATAGAGGAAAGGCTTGAGATAATCACAGTGAATGCAGAAGGAAAAGACAAAGAGATTAAAGCAATTAGAGAGAAGATAATAGATATGGAGGACAGACAAAGACGATCCAACATAAGGATAATTGGTGTCCCTGAAGTAGAGAACCCAACAAATGGAACAGAAAAAGTATTCAAAGATATAATACAAGAAAATTTCCCTGAAATGAAGGAAGAACTGAATCTGCAGATCGAAAGGGCACACCGTGTTCCAGGAAAAATTGATACAGAACGATCGACACCGAGACATATCCTGGTTAAGTTATTGAACTTCAAGGATAAAGAAAGAATTCTTCAGGCATCCAGGCAGAAAAAGCAAGTCACCTACAAGGGGGAAAAAATCAGGCTGGCCTCAGACTTCTCCACAGCAACATTCAATGCCAGAAGACAATGGAGCAATGTCTACAAAGTTCTGAGGGAAAGAAAGTGTGACCCAAGAATNTTATACCCAGCCAAGTTGTCGTTCAAGTATAAAGGCAACAGGCAGACATTCTCAAACATGAAAGAACTCAGGGAATATAGCACCCATGAGCCCTTCTTGAAAAAACTACTTGACGATGAAATCCAGCCAACCAAGAGATGAATCAAAATAAAGAACTCAGGAATGGAGAAGCCGTGGTAAAAGGACTGGTGGTGAGCACTGAATCCATTTAAATATAGAACTAAGACTAAACAACTGTGGGAATTATGGTTACAGAACAGAATGTAAATGTTATAAACCTTGACAANGTAAAAATAATATAANTAACAAAAATCGGGAGGTGGGAGAGGAGAGGTGGGAGGAAGTGTAAGAGTGCTAATTTCCTCATCTTTCATAGCAGGGAGTCAATCGATACTGTCTAAAATTGAAACATGTAGTTTAAAAANATAATGACTCCAACCTCTTAATGTTTTTCATAATCTTTTTTCTTAACCTTAGAGGGATCTTTTAGGAANTAATATCTCTTGTGGTGAAGAAACATTTATCTGAAGTTCAGCAATTCCTTCAGTTTCACTTCAGTTTCTTTTTCTTCTGTTAAATTCAAGTAAAATTAAATTTAATACTTTTNATTTAAAATAGCATGTATAGTATGATCCCATTTTTATAAAATTATTTCTATCTATCCATCCATCCATCCATCTATCCTTCTATCTGTATATATGCATAGAAAGATGTCTGGAATGATGTTCACCAAATGTTAACGATGGTTATTTCTGGGTGGTGGGATTTNGGGTGATTTNTTGCTTTCTTCTTTGTACTTTTCTGTATTGCTTGAATTTTTTATAATGAGCATGTATCATTTTTATAAAAACAATAAAGTNATTATTTTTTTAAAAA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| HAL1b | BPC5 | 1113 | 1142 | + | -43.05 | AGACAAAGAGATTAAAGCAATTAGAGAGAA |
| HAL1b | BPC5 | 2208 | 2237 | - | -43.68 | AGATAGAAGGATAGATGGATGGATGGATGG |
| HAL1b | BPC5 | 2196 | 2225 | - | -44.44 | AGATGGATGGATGGATGGATAGATAGAAAT |
| HAL1b | BPC5 | 2204 | 2233 | - | -44.99 | AGAAGGATAGATGGATGGATGGATGGATAG |
| HAL1b | BPC5 | 1095 | 1124 | + | -45.33 | AGTGAATGCAGAAGGAAAAGACAAAGAGAT |
| HAL1b | BPC5 | 60 | 89 | + | -45.50 | GAAAAGTGGCGGCCAAGGAAAGAAACAGAA |
| HAL1b | BPC5 | 927 | 956 | + | -48.88 | AAAACAACAGAATGAGATGAAAAGGGAGAT |
| HAL1b | BPC5 | 1320 | 1349 | + | -48.89 | AAAAATTGATACAGAACGATCGACACCGAG |
| HAL1b | BPC5 | 1862 | 1891 | + | -49.22 | GAGGTGGGAGAGGAGAGGTGGGAGGAAGTG |
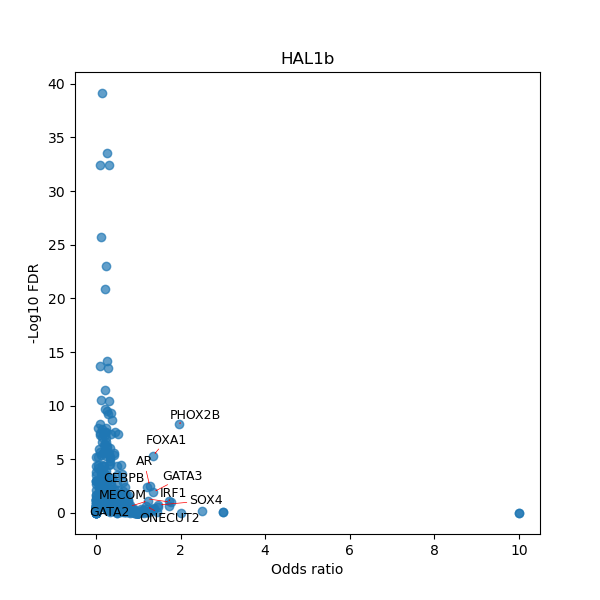
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.