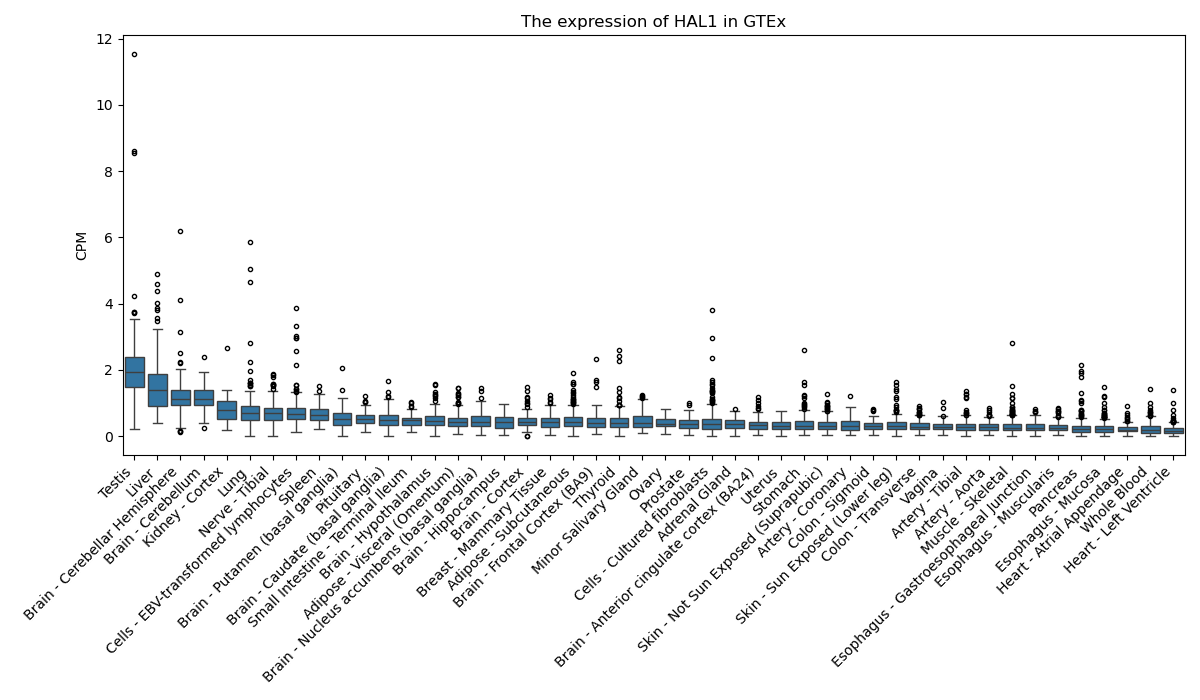
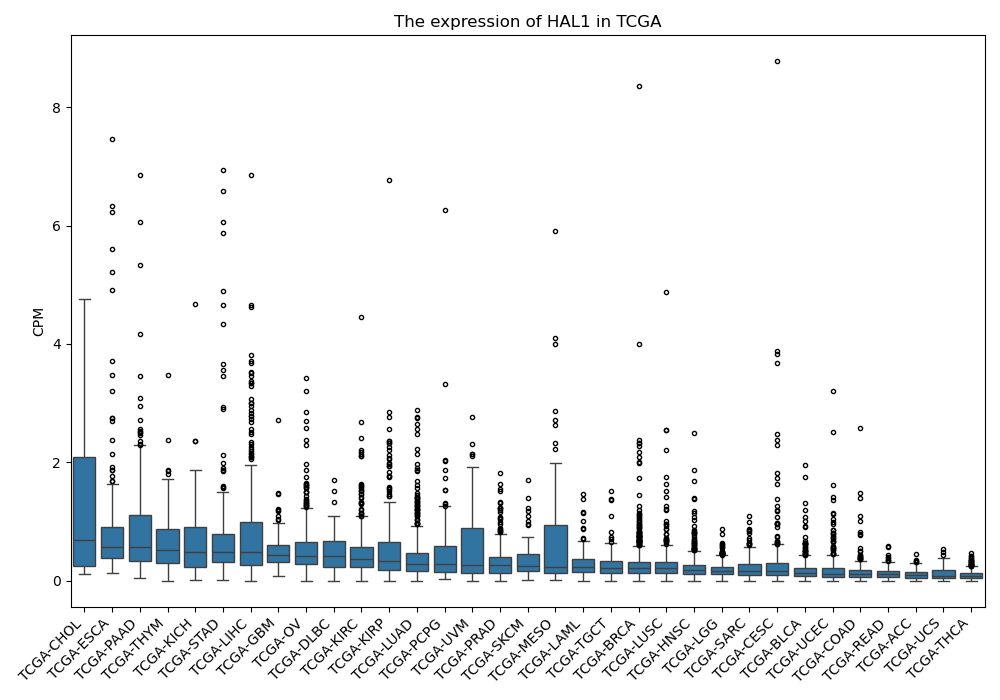
HAL1
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000154 |
|---|---|
| TE superfamily | L1 |
| TE class | LINE |
| Species | Eutheria |
| Length | 2507 |
| Kimura value | 31.19 |
| Tau index | 0.7715 |
| Description | L1 retrotransposon, HAL1 subfamily |
| Comment | HAL1 resembles half of a LINE1 element, as it encoded a protein closely related to LINE1 ORF1, but has no similarity to ORF2. The HAL1 3' end (position ~2300-2510) matches 3' UTRs of the oldest L1 subfamilies. The precise relationship between HAL1 and L1's is unresolved. |
| Sequence |
GACTTCCGGTTAAAGATGGCGGATTGAACACACGCATTTANTTTCGCTCCCTCCCGAAACCCCACTAAAATGACAGTAAAGGAATTTTTTAANAAAGGCATAAACCCACAAGGACAAAGAGAACGGGAGAGGAGACAACAGCAACAAAATTTTGGAAGCTGGAAAGCAGATGGACGAGTGGTAACTGACTTAGCAGACCCGAGAAAGCTGAATCCTAAGCCGGCAGTGGGGAAAGCCGAGAAGCAACCCGATTTACACCGCAGAATCCCCAAAAGGCTCAGGAATTGGCGGCACCAGGTACCTCTGGAAGTGGGGGTGAAGGTGGGGCTAAAAACAGGGAGGATTGGTTGAAAGTCTGTTTAAGAAGCAGTTAGACCCCCAGATCCCCTCCCCCACTCTATNGCAGCCAGGCGACTGCCCCTCCCCCACCCCGGCAGAAGACTGGAGGTTTATTCTCTGGAGAGGGTAAAACAGAGGGTCTCTGGACTGGGGGACACCAGGCACAGTTGAGGGCAGAGGGGTACCGTACTGAAAACAGGGGGATTAAGTGAAAGTNTACATACTGAATGNCGAGAGACCCCCAGCCCTCTTCCCCCACTCGGCTCCCAGAACGCTGGCAGCCAGGCCTATACCCTCCAGGCAGGAGATTGGAAGANTCTTCTCTGGGGAATCTGACCAGCCCAAGAGGAAAGACCTAAAGATACTGACATTNGGGGTTCCCCAAGCGAAATAGCCCAGCCAGATCACCCTACAGTGAAGCCCACAGTCGACAAGCCCCACCCACGCGCNCAGAGCTTCCAATCAGCTTTTTAGTGCCTCACTCTTAAATATGAGCAGACAGCCAAGGATCACCAGACATCTGAGGAAAGCCTCTAACATGAAAGANAGAGACCAAAACAAACAGAAAAAAAAAGCAACTTGGAGGAAACAGAAGACATGCAGAGAGAAGAAAACNTCTTTAAAAANATNATTAATATCCTCAGAGAGATAAGAGAAGATATTGCATCCATGAAACAAGAACAGGATGCTATAAAAAGGAACATTCAGAGAACAAAAAGAAGCTCTTGGAAATTAAAAATATGATAGCAGAAATNAAAACTCAATAGAAGGGTTGGAAGATAAAGTTGAGGAAATCTCCCAGAAAGTAGAACAAAAAGACAAAGAGATGGAAAATAGGAGAGAAAAGATAAGAAAATTAGAGGATCAGTCCAGGAGGTCCAACATCCGANTAATAGGAGTTCCAGAAAGAGAGAACAGAGAAAANGANGGAAGAGAAATTATCAAAGAAATAATNCAAGAAAATTTCCCAGAACTGAAGGACATGAGTTTCCAGATTGAAAGGGCCCACCGAGTGCCCAGCACAATGAATGAAAAAAATAGACCCACACCAAGGCACATCATCGTGAAATTTCAGAACACCGGGGATAAAGAGAAGATCCTAAAAGCTTCCAGAGAGAAAAAACAGGTCACATACAAAGGATCAGGAATCAGAATGGCATCGGACTTCTCAACAGCAACACTGGAAGCTAGAAGACAATGGAGCAATGCCTTCAAAATTCTGAGGGAAAATNATTTCCAACCTAGAATTCTATACCCAGCCAAACTATCAATCAAGTGTGAGGGTAGAATAAAGACATTTTCAGACATGCAAGGTCTCAAAAAATTTACCTCCCATGCACCCTTTCTCAGGAAGCTACTGGAGGATGTGCTCCACCAAAACGAGGGAGTAAACCAAGAAAGAGGAAGACATGGGATCCAGGAAACAGGGGATCCAACACAGGAGAGAGGCGAAGGGAATTCCCAGGATGATGGTGAAGGGAAGTCCCAGGATGACAGCTGTGCAGCAGGCCTAGAGAGCAACCAGTCCAGATTGGAGCAGGAGGACAGAAGGCTCCAGGAGAGATNTCTCCAAGAAAATGAAACTGATAGANTACCTGATGTGTTTGAACATATTGAGAGGAGATTTANACAATTGGCGGAGAGTTTGGGGATGAATTAGTGATAAGTACATAGAAAACTAAGCAAATGAAAAACGAGACAATTATTAACTCCAGGGAAAACAAAAAGTTGTACAAGAAAGGAAANGTAATCATAGTACACTACATGGCTCAGCTGTGAATAATATTTACATAGTCATAATAATGTAAACACTGAATATTGATTTAACCAAAATTATGATATAACTATATTGGGAGGATGGGGGGATGGGAAGTGTGTGTGTGNTGAGGGGGAGGTGTGAAAGAAGAGCTAAATCCTCATCTTCCATAGTAGGAAGTCAATAGATAATGCCTAAAATTGAAAAATCAAGAAATAGCAATATAAGCATGTTATTTAGAAATATGGAGGTAAATACCAGAAGAAACAGCTAAAAGAGTTGAAAGTGGTTGCCTCTGGGGAGGGAGTGAAGATGGAAGAGGGNAGGGATGGGGACTGCTTTTCGTNATAAGCCTTGTAGNACTATTTGACTTTTTAAACTATGTGCATGTATNACTTTGATAAAAATAAAA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| HAL1 | BPC5 | 1150 | 1179 | + | -47.30 | ACAAAAAGACAAAGAGATGGAAAATAGGAG |
| HAL1 | BPC5 | 1162 | 1191 | + | -48.16 | AGAGATGGAAAATAGGAGAGAAAAGATAAG |
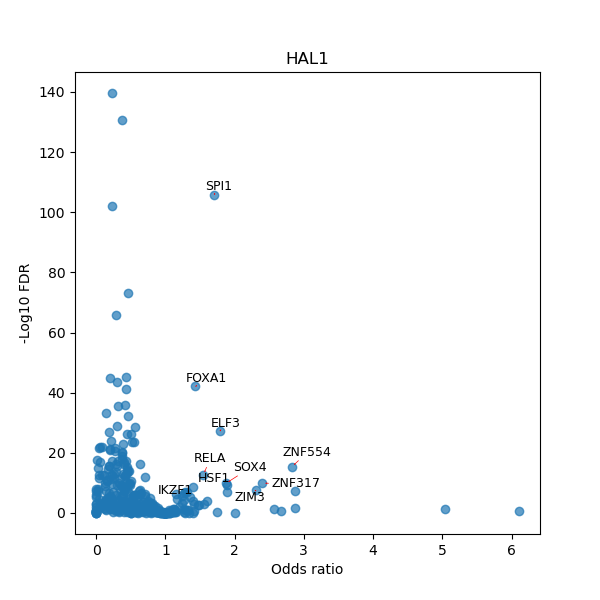
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.