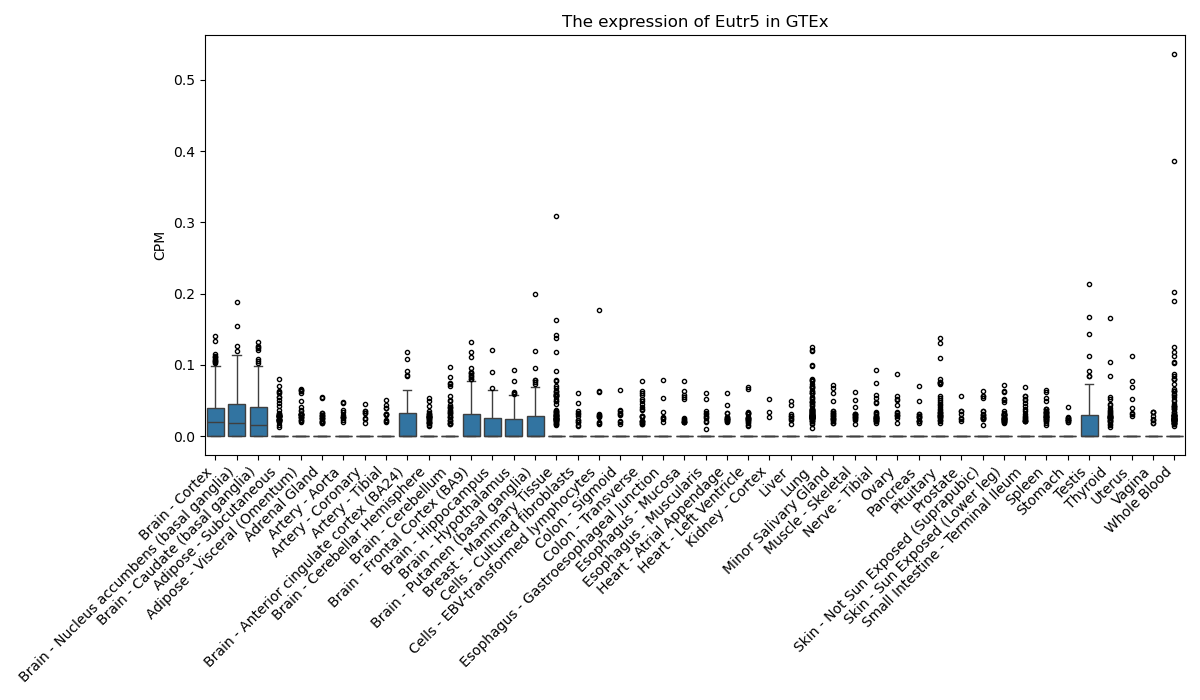
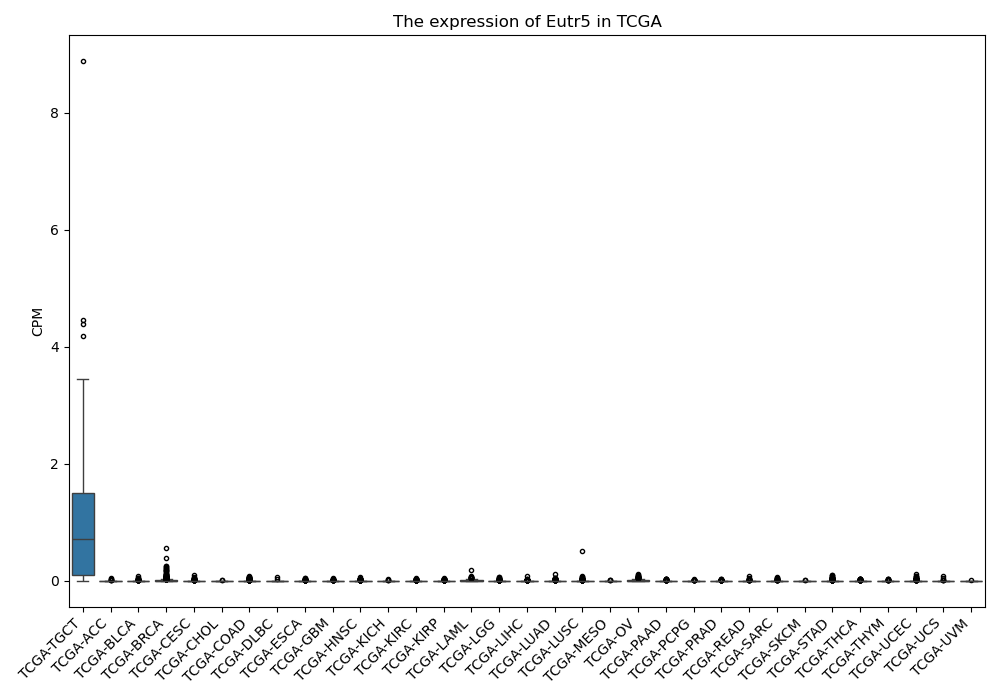
Eutr5
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0001258 |
|---|---|
| TE superfamily | Long_Terminal_Repeat_Element |
| TE class | LTR |
| Species | Eutheria |
| Length | 1130 |
| Kimura value | 34.16 |
| Tau index | 0.9620 |
| Description | Ancient interspersed repetitive sequence from eutherian mammals. |
| Comment | >71% identical to consensus. Origin unknown. More than 200 copies in eutherian mammals. Not present in opossum. ; but it shows no similarity to known SINEs and LINEs. Provisionally guessed LTR because of TG...CA termini but no obvious TSDs nor an obvious (AWTAAA) poly A signal in either orientation. GA-rich 5' end and C-rich satellite-like 3' tail support classification as a SINE. Present at orthologous sites in elephants and humans, absent in opossum. |
| Sequence |
TGTGGCGGGATGAACTGAGATCCTCTTATCCATTCCCCTCCCTCCCGCGGTCAGGGGAGAAACCTGGGGATTAGAGAGAAGCGATTTGGGCATGGCGCCATTCATTGTCAGTTAATGTGAGGAAGCTTTGTAATGTAACCCAGGGCAGGGAAGGGGCAAATGCTCTCTTAGCATAATAAGGCCAGGGTTATTATTTCGCAAGAAATAGGGTCAAAGTTATGTTAATAGGAGGAATATGAATTTCTCATGTAGACCAGACTGGGTGGTCTTCGGAAGGAAATTAATCCTCGCGGTGGCGAGGTGTGTTTGATTTTAGATACTTTAGGAGAAAACCAGCCGCAGCCCGGAGTGGAAGCCCCACTCTGTTCCCACCCCCCTCCCCTNCCCCCAGGACCCAGAGCTCACCCGCCTCCTTTCCTGGGGAGCCAGCGGACCCTGAGACCAGAGCCCACCCCTCCTCTGCAGAGCCAGCAGAANCTGGGCAGAGGAGAAAAGCCACTGACATTCAGCGGACGCCTGAGGCTGCGGAATTTGCAAGACACAAGAAGGGACTAGGGAGACTGAGACTTGATTATTTCTGTAGGGAAGGGTGGCGGATCCCGGGAGAAAGAGGGATTGGATGGGGAAGCAGAAGGAGAAGAGGTTAAAGGGAGAGGGCNGNAGAAATGAGGGAGAAGATAGGAAGCGAGGGAGCGTCCACGTGGGACGCCCTCCGAAGGCCTAGGCAGTGAGGCCTCGTCCCTTGTGTGGTACAAGGGAGCGAGGTAGCTAGACCAGAGNCGCAATCTCCCATCTCCTTCCTCCATCCCGCTTTGGGACAATTACTTGAACCCTTCAGTAAAGAACTGCTTAAACCTCCATCGGAGTCCAGCGTGTATTCGGTTCGGGGAAGACGGGCGAGGCGGTATTCGGANCGGGGAGGGATAACCCCGCTCCNCGGCCATCTGATGGGCATCGGCAGCNCCCTCNCTGTGGACTACTGGGCGGCGAGGAGCATCNACCCGCAGACGACGACCCAGCTGGAGAAACGGTAAGGCCGATCCCCCCGACTTCCCTCCCCCCAGAGAGGCGACTAGAGACATNATCTGTGGGACAGAAAAGGGAGGAGCCCGCCACATGGGTCACATACA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Eutr5 | AT4G12670 | 67 | 95 | - | -6.39 | CCATGCCCAAATCGCTTCTCTCTAATCCC |
| Eutr5 | EWSR1-FLI1 | 401 | 418 | - | -8.42 | GGAAAGGAGGCGGGTGAG |
| Eutr5 | EWSR1-FLI1 | 677 | 694 | + | -9.97 | GATAGGAAGCGAGGGAGC |
| Eutr5 | GLIS1 | 704 | 718 | + | -10.27 | GGACGCCCTCCGAAG |
| Eutr5 | EWSR1-FLI1 | 673 | 690 | + | -11.79 | AGAAGATAGGAAGCGAGG |
| Eutr5 | EWSR1-FLI1 | 580 | 597 | + | -15.50 | GTAGGGAAGGGTGGCGGA |
| Eutr5 | EWSR1-FLI1 | 752 | 769 | + | -15.66 | ACAAGGGAGCGAGGTAGC |
| Eutr5 | DAL81 | 332 | 350 | + | -39.14 | ACCAGCCGCAGCCCGGAGT |
| Eutr5 | BPC5 | 626 | 655 | + | -39.95 | AAGCAGAAGGAGAAGAGGTTAAAGGGAGAG |
| Eutr5 | BPC5 | 595 | 624 | + | -48.40 | GGATCCCGGGAGAAAGAGGGATTGGATGGG |
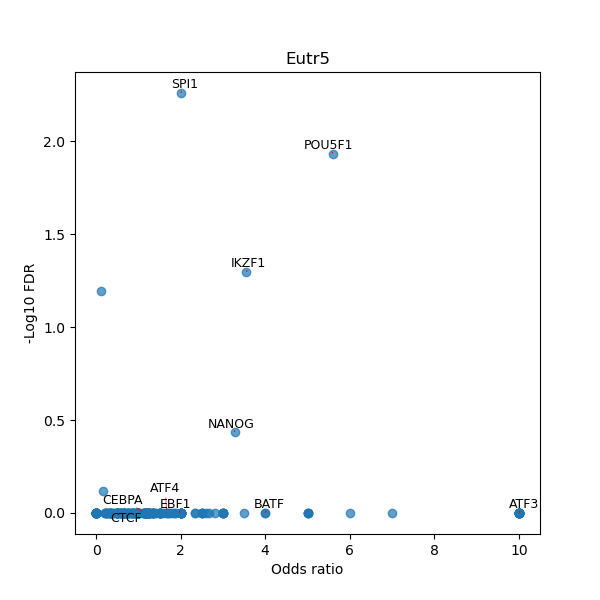
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.