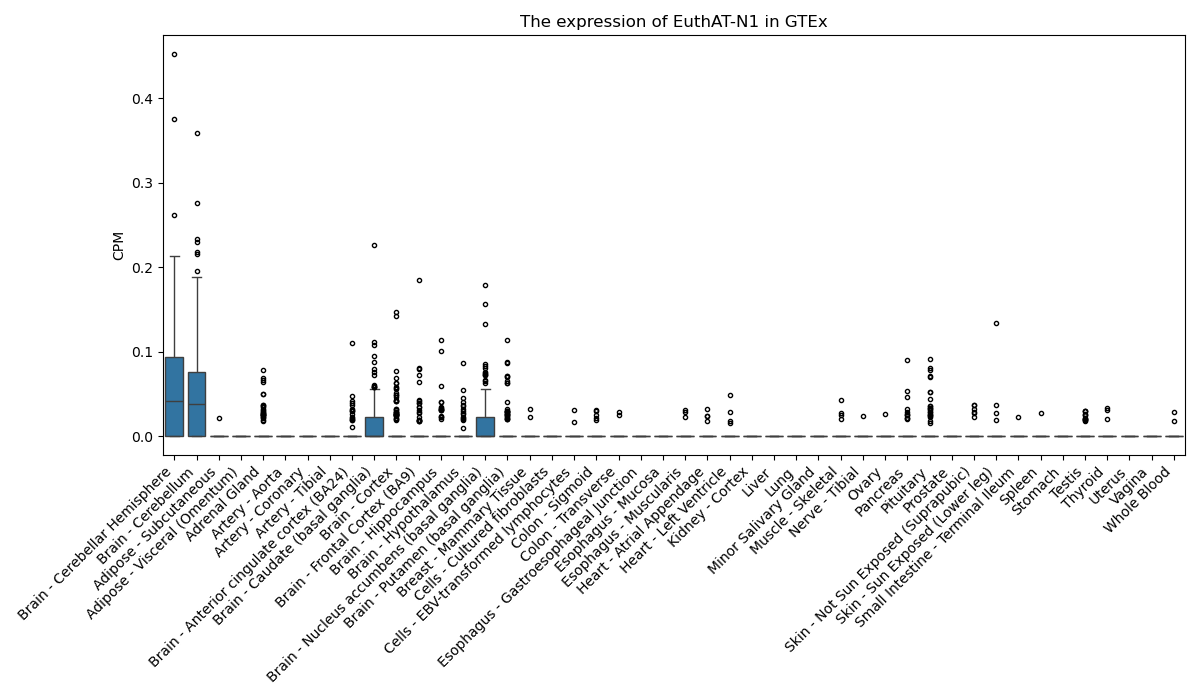
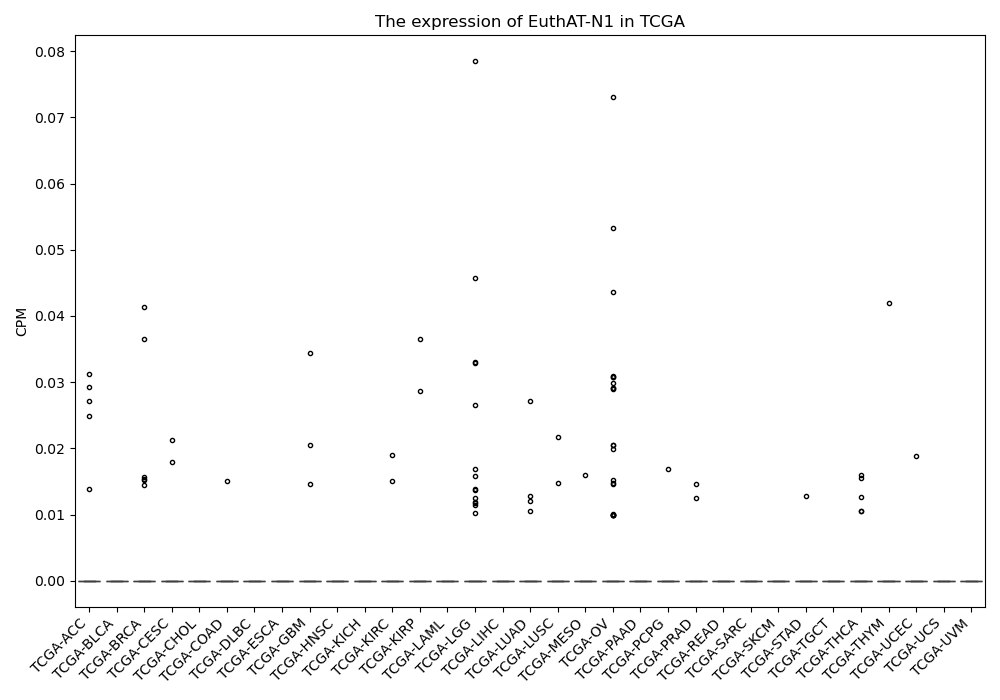
EuthAT-N1
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0001168 |
|---|---|
| TE superfamily | Tip100 |
| TE class | DNA |
| Species | Eutheria |
| Length | 335 |
| Kimura value | 28.51 |
| Tau index | 0.9795 |
| Description | Ancient hAT DNA transposon from eutherian mammals. |
| Comment | 8 b TSDs. Bit improved (original entry 91% similar to pos 1 -325). Very similar to hAT-N1_MD in marsupials, but a different non-autonomous deletion product. At orthologous sites in all eutheria, but apparently absent in marsupials. |
| Sequence |
CAGAGCCGTAACTAGGGCGGGGCGACCGGGGCTTTGCCCCAGGGCACCGAAATTTAGAGGGCGCAAANATTTTAAATAATGATTTTAAAAGATTGTGCAATTTTAAAATTTGTTACATTTACATGTTGACAGCACATGCACTCCTTCAGCAGCGTAGAAACAACTGAGCTTAGCACCTAGTTAGCAAGAATAATTAGTTAAAATAGGAAGCTACCAGATGGCGCGCATTGTTAATAACACCCATCTGTGAGCCACATCATGAATTACAAAGTTGATTTGAGGGCGCGTTTCNTGCTACTTGCCCCGGGCACCAAAATTGCCAGTTACGGCTCTGC
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| EuthAT-N1 | ATF6 | 250 | 262 | - | 6.18 | TCATGATGTGGCT |
| EuthAT-N1 | BZIP44 | 250 | 262 | - | 6.17 | TCATGATGTGGCT |
| EuthAT-N1 | ZNF8 | 105 | 124 | - | 6.09 | ATGTAAATGTAACAAATTTT |
| EuthAT-N1 | KLF13 | 233 | 249 | + | 4.98 | AATAACACCCATCTGTG |
| EuthAT-N1 | DOF4.3 | 267 | 279 | + | 4.02 | CAAAGTTGATTTG |
| EuthAT-N1 | phol | 214 | 223 | - | 3.28 | CGCCATCTGG |
| EuthAT-N1 | ESR1 | 14 | 28 | + | 2.68 | AGGGCGGGGCGACCG |
| EuthAT-N1 | ESR1 | 14 | 28 | - | 1.74 | CGGTCGCCCCGCCCT |
| EuthAT-N1 | ZSCAN16 | 121 | 138 | + | -20.43 | ACATGTTGACAGCACATG |
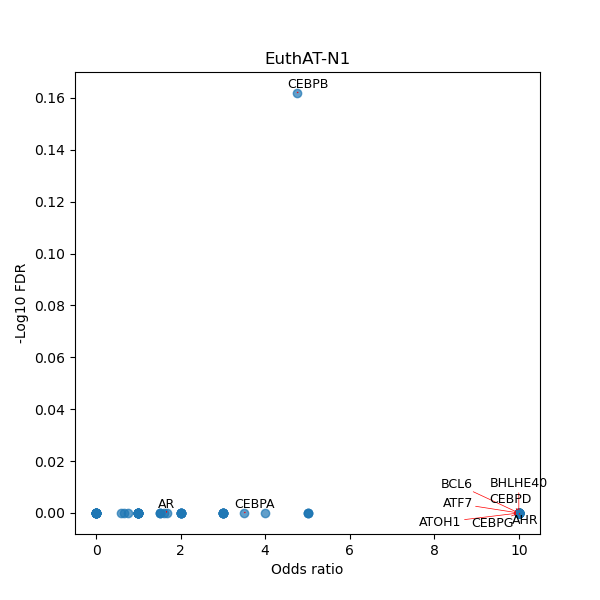
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.