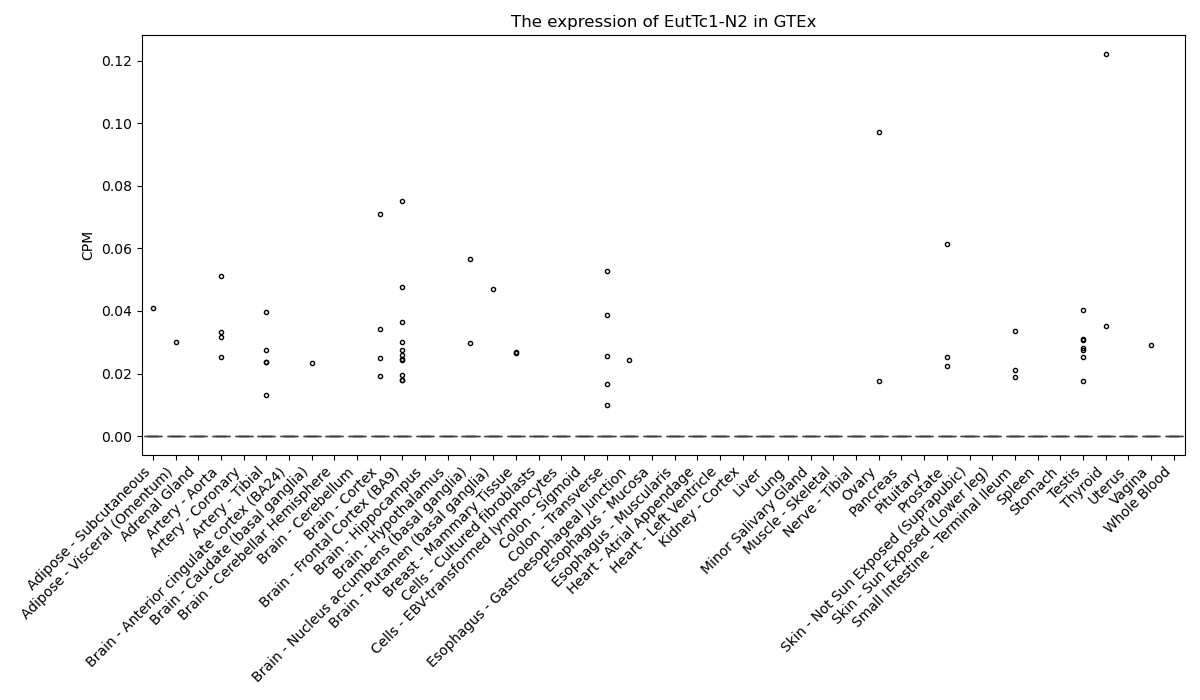
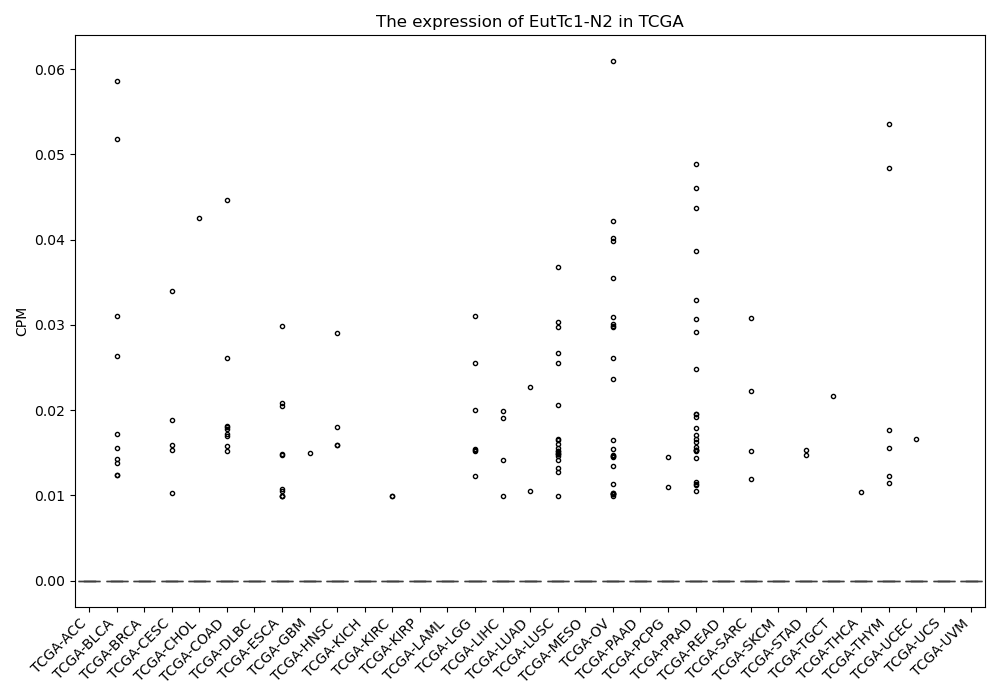
EutTc1-N2
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0001231 |
|---|---|
| TE superfamily | Tc1 |
| TE class | DNA |
| Species | Eutheria |
| Length | 378 |
| Kimura value | 20.04 |
| Tau index | 0.0000 |
| Description | TcMar-Tc2 DNA transposon, EutTc1-N2 subfamily |
| Comment | Hairpin with just pos 177-204 as a unique loop (the first/final 168 bp are basically the same as in EutTc1-N1). Present at orthologous sites in all eutheria, absent from marsupials. |
| Sequence |
CAGGGTGTCCCAAAAGTCTTAGTGCAGTTTTAAGCTTTAATAACTTCAGAAGTATAAATGCTACAAACTTACAAAAAACATCATTTGAAAGTTTAATTATTTAAATTTCTTTTACACTTATTTAGTTTTGTGAATTTTGAATAATAAATTTTTAATTTTAATTTTTTTGTTTCAGTCCCTCTGATTGAAGATGGCAAACGTTGACTGAAACAAAAAATTAAAATTAAAAATTTATTATTCAAAATTCACAAAACTAAATAAGTGTAAAAGAAATTTAAATAATTAAACTTTCAAATGATGTTTTTTGTAAGTTTGTAGCATTTATACTTCTGAAGTTATTAAAGCTTAAAACTGCACTAAGACTTTTGGGACACCCTG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| EutTc1-N2 | lin-54 | 269 | 281 | + | 14.07 | AGAAATTTAAATA |
| EutTc1-N2 | lin-54 | 98 | 110 | - | 14.07 | AGAAATTTAAATA |
| EutTc1-N2 | DOF3.6 | 152 | 172 | + | 13.80 | TTAATTTTAATTTTTTTGTTT |
| EutTc1-N2 | WRKY25 | 199 | 206 | - | 13.77 | AGTCAACG |
| EutTc1-N2 | TCX6 | 273 | 287 | + | 13.67 | ATTTAAATAATTAAA |
| EutTc1-N2 | TCX6 | 92 | 106 | - | 13.67 | ATTTAAATAATTAAA |
| EutTc1-N2 | SOL1 | 273 | 287 | - | 13.66 | TTTAATTATTTAAAT |
| EutTc1-N2 | SOL1 | 92 | 106 | + | 13.66 | TTTAATTATTTAAAT |
| EutTc1-N2 | WRKY30 | 199 | 207 | - | 13.65 | CAGTCAACG |
| EutTc1-N2 | TCX6 | 266 | 280 | - | 13.64 | ATTTAAATTTCTTTT |
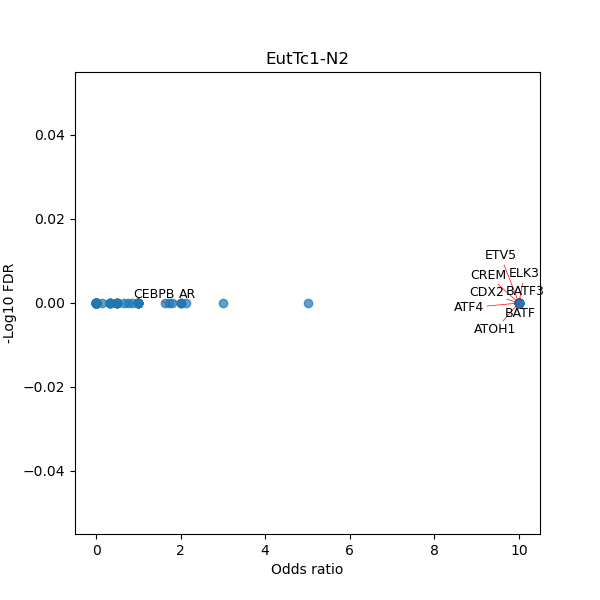
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.