Eulor11
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000123 |
|---|---|
| TE superfamily | Transposase |
| TE class | DNA |
| Species | Amniota |
| Length | 552 |
| Kimura value | 32.76 |
| Tau index | 0.0000 |
| Description | Eulor11 (Euteleostomi-conserved low frequency repeat 11) |
| Comment | Putative DNA transposon (assignment unclear). Identified in both mammals and birds. Reconstructed from chicken DNA. Like most Eulor-type repeats, this repeat has a secondary structure (imperfect hairpin) followed by a 50 bp "tail." The tail region is less conserved in mammals than the secondary structure. Over 300 copies in alligator. Termini uncertain; no similar TEs. |
| Sequence |
TGCTCTCANTACGATAGCTTTCAAAATCTAATTTTGCCCCCTTTTAAATGTGCTAAATATAAATGCAGAAACAAAGAAATATACATTGATAAGTAAAGAAGCACAGGTGTACCTTGCAGGCAGTATAAAAAAAGTCCTTTAAGAGGTCTTGAACCAGTGTCCCTACGTGCTTTATACTGCAGCCCAATGTGCAGCCACTAGACCACCCCACCACAGGTAAGAAATGAGAAATTTTGAGGGCTATTGAACACTGCGAATTTTCACAGTGGATCAGCACAAAGTTATTTAGCACAGGTGTTTCTGTAATTGTGATACATTGGGAAAATTCACAGTGTTCAATGGCCCTCAAACTCACACTTCCACCTGCGTGGTGCGGTGGTCTAGTGTTAGTACACTGGGCTGTAATATAAAACATGTAGGAATACTGGCTGGCTCAAGACCTGCTAAAGGTTTTTCCTTATACCGTCCGTGCTTCTTTGTTCGTCTGGATTTCTTTGTTTCTGCATTTATATTTAGCACATTTANAAAAAGGGGCGAAACAGATTTTNCAGG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Eulor11 | BPC5 | 57 | 86 | + | -44.27 | ATATAAATGCAGAAACAAAGAAATATACAT |
| Eulor11 | BPC5 | 61 | 90 | + | -49.23 | AAATGCAGAAACAAAGAAATATACATTGAT |
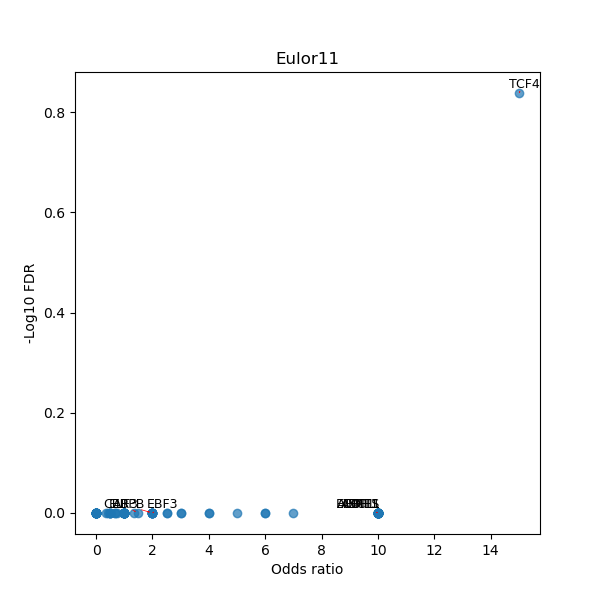
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.