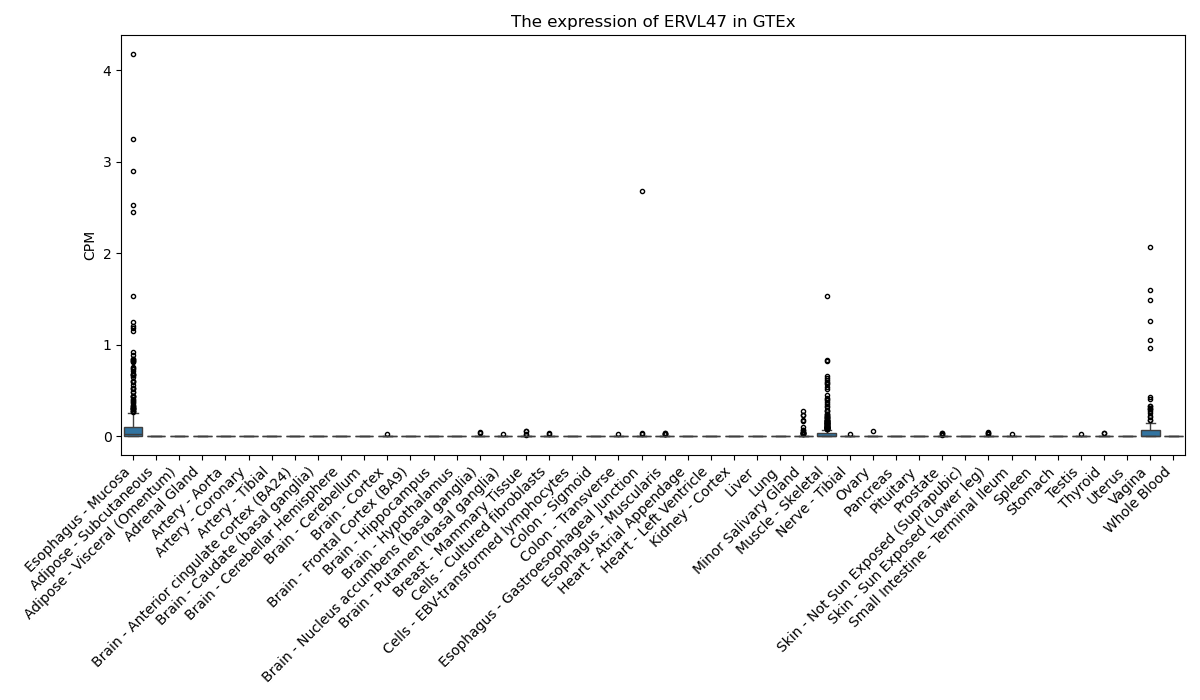
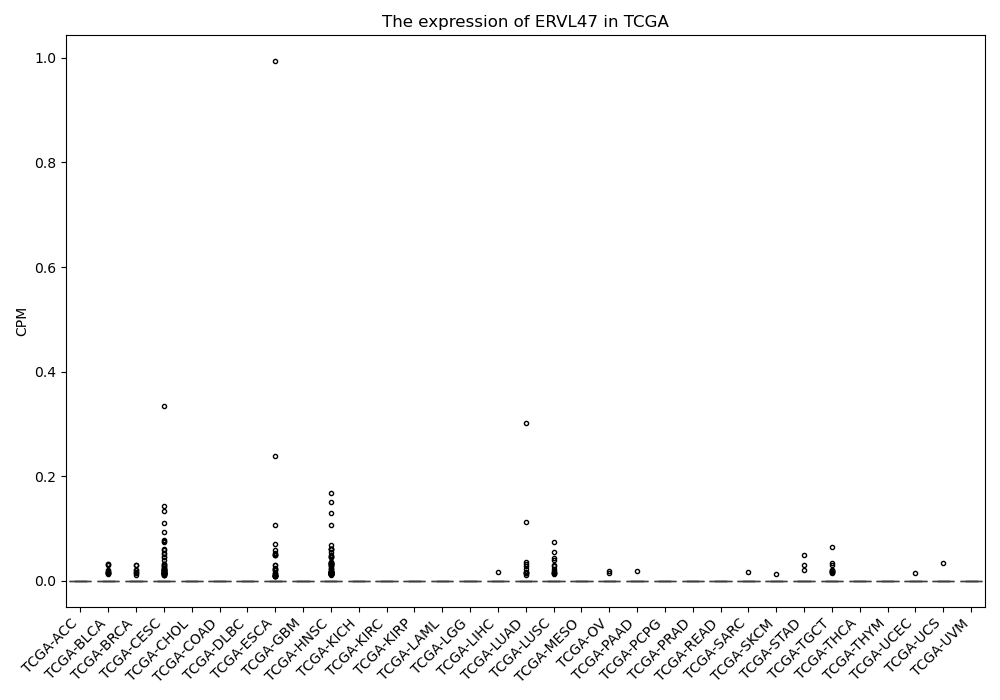
ERVL47
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000119 |
|---|---|
| TE superfamily | ERV3 |
| TE class | LTR |
| Species | Primates |
| Length | 5246 |
| Kimura value | 18.41 |
| Tau index | 1.0000 |
| Description | ERVL endogenous retrovirus, ERVL47 subfamily |
| Comment | Associated with LTR47B2 LTRs. ORF coordinates are roughly: Gag (234-1559), Pol (1560-4955). |
| Sequence |
AATGGCGAGCCAGCCAGGAGGAGCCNGGGAACCAAAGTATGGGCAAGGGGAAGGAAGCATCCGTGGGGGAAATCCCGGGGCGGCCCTCCTCATCTATGTGGGGTGGTGTTGCCCATTTGCTAGATGCCTGTGGACCACCGCGTGAGTACGGGGACGCCCCGAAAACGCCGGAGGGTCTAGAANGCTTGTTAGTAGAAAGCCGTATGACTCACTGTGGTAAGGAAGGTCGCNAGGCGGCGGCAACAGTGGGTTNGCCGCTGTTATGGNCTGTGCGAGTGGCCACAGAAGCGCAGGTAGCAGCTGAGGCGAGGGTAGGAAAGCTAGAGAAAGAATTACAGTTAGAAAGGGATATGAGAACTTCCACGTCTGTGTTAGCGTCTGGTTTGGTGGATAAGCTCGAAACTCAGGAGGCACAGTTGGAAACCGTAGCCTGCCGCTTTGTGCGGCTGGGAGGGAGGAAGCTGCGCCGGCCGAAAGTGCGGGCTGTCCTTGCCAAACCGGACTGGGATGCNAANACCTGGAATTCTTGGGAACCAACTAGTGAGTCAGATGANGACATAGAGGTTANCTCAGAGGAGGAGGATAATTATCCTCCCCTTTTGAGAGCGAGACCGCTCATGCAGCGGAAAGCAAAAGCCCAACACATGCAGCCGGGCAATAACGGACAGCCTTTTCAGGAAACCTTAACTGTCCGAGAGTACACCTCGGCAGGATTATTGGACATAGCAAAAACTTTTAAACAGCTGCCGAGAGAAAGCTTAGCCACATGGATGGTGCGGCTATGGGACACCGGTGGGGATGGCATCTCCCTAGCAGGGAGTGAAGCTGAAAAAATGAGTAATATAACCACCCATCCNGCTTTGAGACAGCATTTGCATAATGCTAGAACTGTCGAGGGNAATCATAGCCTTATGGACTGGATTATCCTGGCCATGAGAGAGGCTTGGCCNAATGAAGGGGACTTCCCGGGTCAGACCCCGGCATGGCGATCTCTGGAGGAAGCGCAGAGCGATTTANGGGAGTTGGGCATGCGNCAGGCCATCTATGCCCAGCAATTTGNAGGACCAGATAAGGCTGTGTTTACCGTGGGCATGAAAAATAAGTTGTTACAAAGTGCCCCCCGGGAGTGGCATGGCCCTCTCATATCCCTCTTGAGCCCCCTTGTGGGACAAGATGTATATGACGTGGGGGAAGCCATNNCTGATCTTGGAGAAACTGAAAAGGGAAGAGATAAGGTCCGACTGGTTACTAAAGGAAAAGGGAAAAAGGGAGAAACCAAGTCTGCTGGACTGAAAAAGGGAGGTCAAAAAGGCCCAGTTAGGATTACCAGGAAACAAATGTGGTATGATCTGATCTCAGCTGGGGTAGACAAAGAGAAAATAGACCGGCAACCCAATGCCATATTAGTGGGCCTTTGGAAAGACCTGACTCCCGATCAACAGTTTAGACCCCTTCCTAGTGCCCCACCAGAAGAGGGAGAAGGAGAAGAAGAAACTCCGCGTAAAAGAACCCCAATCTCTGCGTTCCAGGGGTGGACTCCTCCCCAGCCTAGAGACTAGGGGTGGGGCCAAGGTTGCCTCCGTGTANGAGCAATAGGGGGCGACCAGAGGCCCCATGTGGAGCTCACAATCTATTGGAGTCCAAAAAACAAGCAGAGAACCTTAGCTTTAGTGGGCACAGGTGCAGAATGCACCTTAATTCATGGAAATCCAGAGAGACACCCTGGTAAGTGGGCAGCTATAGATGGTTACGGGGGGCGAACAATCCGAGTGAAACAAACTCCTCTCCTCCTTGGTNTTGGGCGGAGTCCCCCCGCTTACTNTACTGTCTTTATCTCACCTATTCCAGAAAACATCTTGGGTATGGATGTCCTTTTAGGATGCACTTTACAAACATCTGTGGGGGAATTCCACCTATGAGTTTGGGCGGTAAAGGCTATTTTAAGAGGGGAAGCAAAATGGGAACCTGTACACCTCCCTCCCCCACGGCGTATTGTTAATGTGAAACAATACCATCTTCCTGGTGGGATAGAAGAAATCACAGCCACCATACAGGAACTGGCCAAAGTTAATATTATNCGGCCAGCCCAGAGTCCCTTCAACAGTCCTGTATGGCCGGTAAGAAAACCTGATGGCACCTGGCACATGACAGTAGACTACCGGGAACTAAATAAGGTGGTTCCCGAGATACACGCTGCTGTGCCTAATATAACTCAAGTGATAGAGCAAATAATACAAAATATAGGCACTTATCATGCTGTGTTAGATTTGGCTAATGCCTTCTTCAGCATCCCTTTACACCCTGACTCGCAGGACCAATTTGCTTTTACTTGGAATGGCCAACAATGGACATTCCAAGTGTTGCCCCAGGGNTATCTACANAGCCCCACTATTTGTCATGGAATGATTGCTAGAGATTTAACTTTATGTCCACTGCCACCTGCTGTTAAACAGTTTCATTACATTGATGATATTATGCTAACCTCTGAAGACTTGTCATTGCTACAGCAACACCTTGATGCATTGTGCACCCTTCTCCAATCCAGAGGATGGGCCATCAACCCGCAAAAGATACAAGGCCCAGGACCGGCTGTAAAGTTCCTAGGGGTCACTTGGTCGGGTAAGACACGCCTTATCCCAGGCATAGTCATTGACAAAATACAACAATTTTCCATGCCTAAAACAGTTAAACAGTTACAAAGTTTCCTAGGTCTTTTGGGATATTGGCGGGCTTTTATTCCACATTTAGCTCAATGTTTGCGTCCCCTATACCGACTAGTAAAGAAGGGATCTAGTTGGTGCTGGGATAAAGAACAAGAGGAAGCATTTGAGAAGGCTAAANTATTAGTGGCTCGGGCACAAGCCTTAGGTTCCCCCCTTCCCGGGNTACCGNTNTCTTTGGATGTGACCATAAGCCCTGAGGGGACCAGCTGGGCCCTCGGGCAAGTCCAGCATGGGAAAGCGGTTCCCCTAGGATTCTGGTCACANCTATGGAAGGGCGCTGAAACCCGCTATTCCCCGATTGAACAACGGGTCCTGGGGGTATANAAGGCCTTGCGGCAAGTTGAACCCGTAACTGCCGCTTTNCCGGTAACAGNGAAAACGGGTCTCCCTATNAAGGGCTGGANAGAAGGGTTGTTTNCCAGGCCTGNNTCAGCTATTGCCCAGGCCTCCACTTTACAAAANTGGCATGCANACCTGCAACAACGTAGCGCCCTCTCCACGAGTCCCTTGGGAGATGAACTGCATGCTNTCTTAGGGCCAGTACACTATGAGACCAGTGCTGCCCCTATTGTGGAGCCCCCACGGGGGATGCCTCCGATGNTACACGAAGGCACGGCCCCCATTCCTGAAAACGCTTGGTACTCGGATGGGTNNAGCCGAGGTAACCCTTGTGTATGGACGGCAGTAGCTGTACAACCGCAGACAGATACTATCTGGTTTGAGATGGGAATGCAGCAAAGCAGTCAATGGGCAGAACTCCGAGCTGCATGGTTGGTTTGTACCCATGAGCCATGGCCTATAGTTCTCTGTACAGATAGTTGGGCAGTATTTAAGGGTCTTACAACTTGGCTTGCCCAGTGGGCCCGGGATGACTGGCATGTACTTTAAAAACCCTTATGGGGAGCTGCCATGTGGAAAGACATTTGGGAAAGGCTACAAGAACCCACTGCGAGCCTAATTGTGTATCATGTTTCAGCACACTGGTCAGATTCACCTCCCGGTAACATGGAGGCTGATACCCTAGCAAAAATTAGAACACTGGCTCCCTCGCAATCNTCTGAGCTAGCTGATTGGGTACATAAACACAGTGGGCATCGCAGTGCACGAGTGGGCTGGCAAATAGCAAAGGGAGCAGGATTGCCCCTCCGCTATGCAGATTTAGCGGCGGCAGTAACAAACTGCTTAGTTTGCTCCCGCCTGCGCCCCCGCCGCATCCCACATACACCTGGACACATACATAAGACAGCCGCCCCTGTGAGAGACTGGCAGATAGACTACATCGGACCCCTGCCAGTAAGCTTGGGACAAAAGTATGCACTAACATGTGTAGACACTGCCACGGGATTGTTGCAGGCCTTCCCTTGCAAAAGGGCAAACCAAACAGCCACCATTAAGGGCTTGGAGCAACTCAGTGTCATGTATGGATACCCTCGATGCATTGATAGCGACCGAGGCACGCATTTCACTGGACATGATGTCCAAGATTGGGCACATGAAAAGGATATAGACCGGAGATTTCACTTGCCATATAATCCCCAAGCGGCAGGGTTGATTGAAAGGAAAAATGGCATTTTGAAGGCACAACTGCGAGCACTTTCACAATCCAATACCTTGCATGGGTGGGCGAAGGTTTTGCCTCAAGCCATTAGAAACCTTAATTCGGTTGAGACAAATACGGGGCTGGCACCATACCAATGACTCGGGACCACCGCAGAGGAGGGTCCATTAACCATAGTTGTAAAGAAAGTCCGACCAGACGCATTTCTACCGGAGCTGATAAAAGGCCAATGGCAAATGTTATTTGGGACTCCCCAAGACCTTGAGCCAGGGGAGGGGACACTTGAATGGGGGTTGGACTGGCAACTTCCCCCAGGTTGGATAGGGTATTTCTTGCCAGAGAGCGAGGAATTCCCCGGCCAACTAAAGTGGTCTCCGTTGATCCTGTTGGAGTCTGGGCCAAAACGCTCCACATACCAATACACTGGAACACGGCCCCTTTTAAAGGGCACTCTGGTCGGCCGTTTGACATGGTCCTTTGCTGCCCCTGTAACCTTNACAGATAATACCGGCACCTTCGCCCTTAGGCAACATGTTTGGTATGCACCCCCAGCCCATAATCCTTNGGCTGCCTATGTCCTAACTAACAGAGATGAAGCTACCACAGTCATTTTGCTTGATGGGGAAGAACTGCCCCGCCAAGTACCTACTAAACACTTGTATTTCCGCCCATAGTCTTCTGTTCCTGTTGCTGCTCTGCCCTACAAAAGCCTCTTGGTTTGGATCCTGGGGAACATGGTGGCAAAAGGCATTATTAATTCTGTGTCTTATACTTGGCATAGGTATCATTGCCTGTTGTTGTCTGTATTGTTGCTGCGGCCTCTGCTTACAAGTAGAAAACAAATTGATGCAACGTGTCACCCACGCCATGAAATGACTGCAGCACCCCTCNCCTCTAAAGGCTCAGGGACCATCGCGGAAGAGGNGGGCGCGTGAGATTGTAAGAGCCGGATTAGAGGGGTGGAG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| ERVL47 | BPC5 | 323 | 352 | + | -48.98 | AGAGAAAGAATTACAGTTAGAAAGGGATAT |
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.