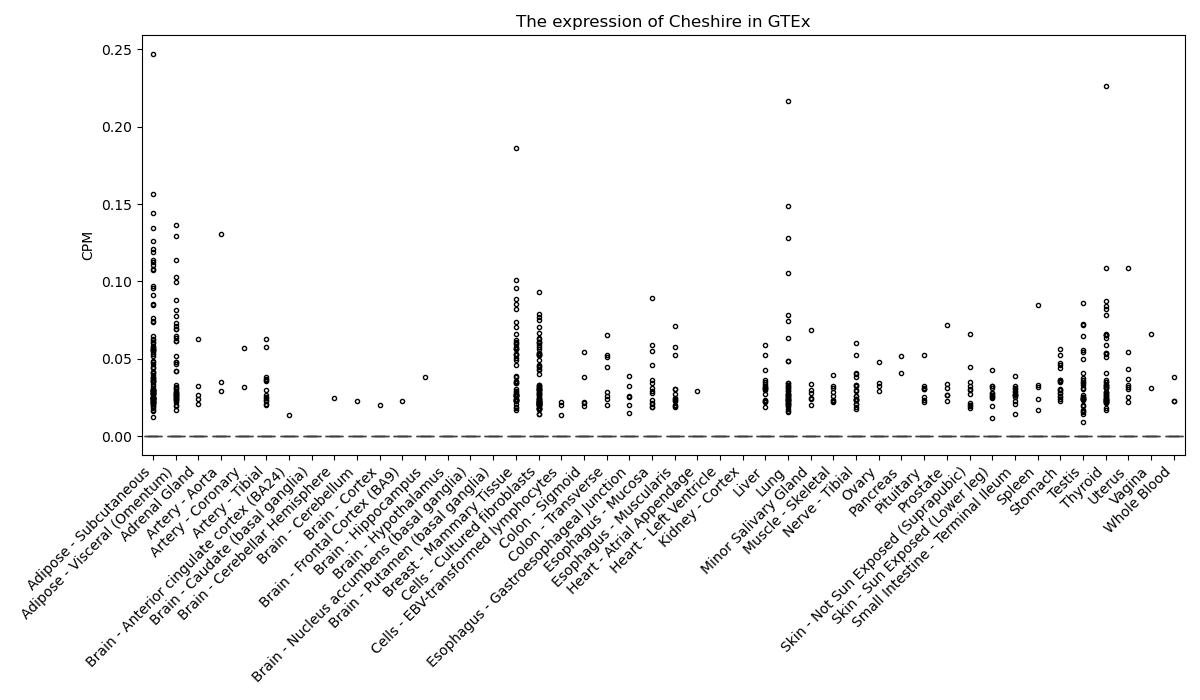
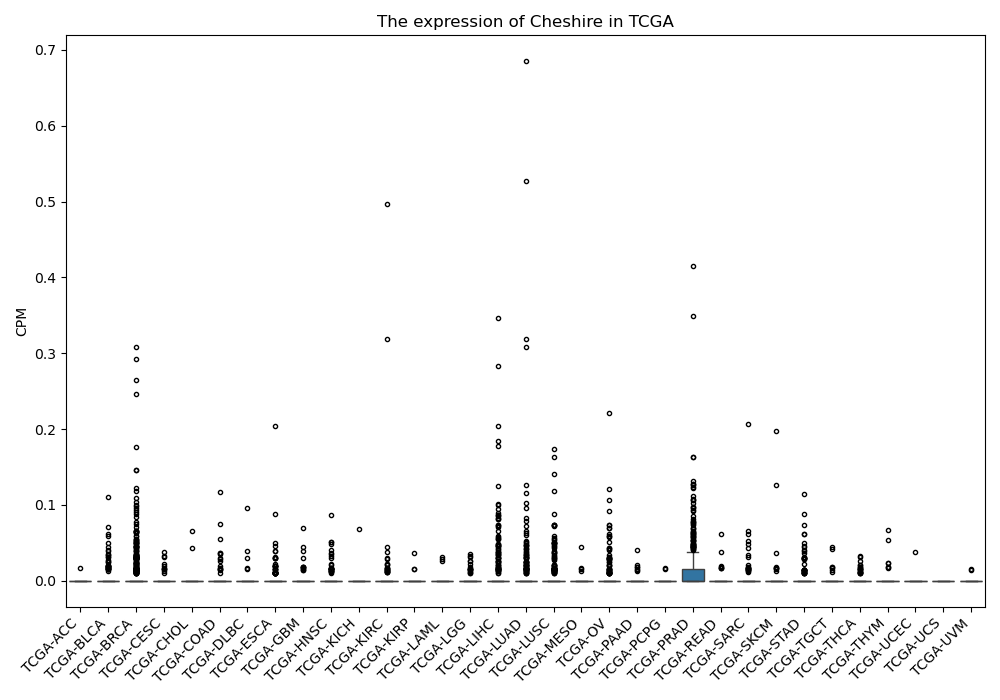
Cheshire
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000109 |
|---|---|
| TE superfamily | Charlie |
| TE class | DNA |
| Species | Eutheria |
| Length | 2420 |
| Kimura value | 18.40 |
| Tau index | 0.0000 |
| Description | hAT-Charlie DNA transposon, Chesire subfamily (autonomous) |
| Comment | 16bp TIR, 8bp TSD. |
| Sequence |
CAGGGGTCGGCAAACTACGGCCCGCGGGCCAAATCCGGCCCGCCGCCTGTTTTTGTACGGCCCGCGAGCTAAGAATGGTTTTTACATTTTTAAATGGTTGAAAAAAAAATCAAAAGAAGAATAATATTTCGTGACACGTGAAAATTATATGAAATTCAAATTTCAGTGTCCATAAATAAAGTTTTATTGGAACACAGCCACGCTCATTCGTTTACGTATTGTCTATGGCTGCTTTCGCGCTACAACGGCAGAGTTGAGTAGTTGCGACAGAGACCGTATGTGGCGCTCTCATCGCTTCGCACTGCTGCTCGGCGCACTACAAATCGCAGTGACGCAGTTATAACTCGACAGCGTTTCGAGTGCCACGCGTATCGCCGTACCGCGACATTTTTTTTTATTACCAGTGCATACCCATCATGTCAAAACAAGAAAAGAAGAGAAAAGTGGACTTCGAATGTCGCGCTTTTAAGGCACAGTGGAGTGTGGATTATTTTGTTATCGAATTAGATGGCAAAGCATTGTGTTTATTATGCAATGACACTATAGCTGTGCTAAAAGAATACAATATACGTCGACATTACCAGACTAAGCACTCATCACAATATTCCCAACTCACAGGAAAGCAACGGTCAGAAAAATTAGAAAATTTAAAACGGAATATCTCATCACAGCAGAATTTCTTCACAAAAATAAAAAATGAAAATGAGGCTGCAACCAAAGTAAGTTTCCGAGTGGCTCATTTGTTAGCCAAGCAAGGAAAGCCGTTTACCGATGGTGAGTTAATTAAATCGTGTTTGATTGCAGCAGCCGAAGAAATGTGTCCAGAGAAAATAAACTTGTTTAAGACTATTAGCCTTTCGGCGAGAACAGTTGCTCGAAGAGTTGAGGACATTGGGAGCAACATCAATAGTCAATTAAAAAACAAGGCAAATGATTTCGAGTGGTTTTCCTTGGCTCTTGATGAGTCGACAGATGTTACCGATACTGCTCAGTTGTTTATTCGAGGAGTCAATGCCGAGTTTGAAGTGACTGAAGAATTAGCCTCTATGAATAGTCTGCGTGGAACAACTACAGGCGAGAATATTTTCAAAGAAGTTGAGAAAACACTAATTCAGTACAACCTGAAGTGGAATCTGCTAAGATGTGTTACAACTGATGGTGGTAAAAATATGTGTGGAGCAGAAAAAGGCTTAGTTGGACAAATTTACAAAGCTTGTGAAAATGTAAGGTGTTTAAAGCCTATGGTTATTCATTGTATTATTCATCAGCAGGTACTTTGCGGAAAATATTTGAATCTATCATGTGTTATTGAACCAGTAGTGTCAACGGTGAACTTCATTCGCTCTCGTGGACTTAACCATCGTCAGTTCCGTGAATTTTTGTCAGAAATAGAAGCTGAATATCCTGACTTGCCCTACCACACAGCAGTTCGATGGCTTAGCAGTGGTAAAGTTTTATTGCGATTTTTTGAGCTCAGGGCCGAGATTGAAATTTTTCTGAATGAGAAGAACCGCCCTCAACCACTATTATCGAACACTGAATGGCTTTGGAAATTAGCTTTTGCTGCAGACTTGATAATGTTTCTTAATGAATTCAACCTAAAATTACAAGGCAAAACAGCGCTTATATGCGAAACTTATACTGCGGTAAAGTCATTTCGATGACAACAACTATTGTTTGAATCACAAGTAATGTCAAGCTGCTTTATACACTTCCCGTGCTGTCAAAAGTTAAAACAAGAAGCGAGATCTCCATTCCCACACAAATTTGCAGCGGATATATTTTCCGAGCTCAAACTACAGTTCCAGCAGCGTTTTTCGGACCTCGATGCAAGTGCAAAGGAAATTTCCATATTTCAAAATCCATTTAACTGTGCAATTGAGGAGCTTCCACCTAACCTTCAATTGGAAGTGATTAATCTGCAATGTAATGACATGCTAAAAGGCAAATATCAAGAGAAGAATCTAATAGAATTCTATAAATGCCTTCCAAGCGATGAATATGCTCAATTAAAATCATATGCTCGTGGATTGATATCAGTATTTGGCAGTACCTATCTGTGTGAAAAGACATTTTCAAAGATGAAATACGTAAAATCTCATTACAGATCAGCATTAACAGATGAACATTTGCAATCGATTTTGATGATAGGGAACACTAACTTTGAACCCCAATTAAGCGAAATGTTATCCCCCCAAAAAGAATTCCATTCTTCTCATTAGTAGACCTGTATTACAAAAAATTGTACTCAATTATTATTATTATTATATTTTGAATTTCGTCAATAAAAATTTTGTGGAAATTTGTTTTCTCTCTTGTTATATAAGTACCTACATAATATCCTCGATTTTGCCTCTTGGCCCGCAAAGCCTAAAATATTTACTATCTGGCCCTTTACAGAAAAAGTTTGCCGACCCCTG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Cheshire | GT-3a | 135 | 148 | + | 15.34 | CACGTGAAAATTAT |
| Cheshire | Zm00001d052229 | 38 | 47 | + | 15.25 | GCCCGCCGCC |
| Cheshire | GRF4 | 801 | 808 | + | 15.23 | GCAGCAGC |
| Cheshire | CRF2 | 40 | 47 | + | 15.20 | CCGCCGCC |
| Cheshire | dl | 941 | 950 | + | 15.17 | GTGGTTTTCC |
| Cheshire | MYB3R5 | 624 | 637 | - | 15.17 | TTTTCTGACCGTTG |
| Cheshire | ZNF75A | 2059 | 2070 | - | 15.16 | TCTTTTCACACA |
| Cheshire | daf-12 | 192 | 205 | - | 15.04 | GAGCGTGGCTGTGT |
| Cheshire | FOXD3 | 824 | 837 | + | 14.96 | AGAGAAAATAAACT |
| Cheshire | CDX4 | 1453 | 1461 | - | 14.91 | GCAATAAAA |
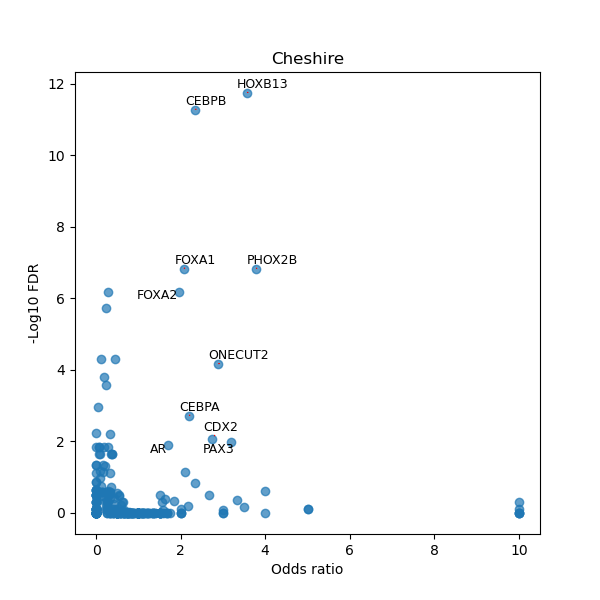
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.