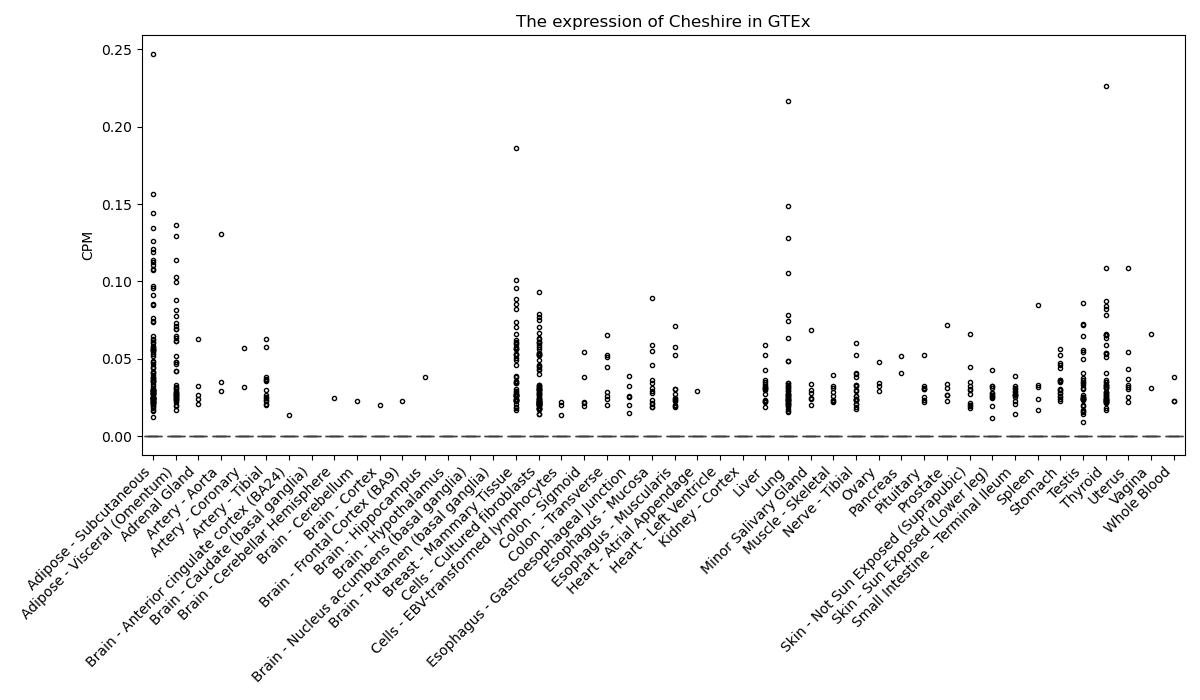
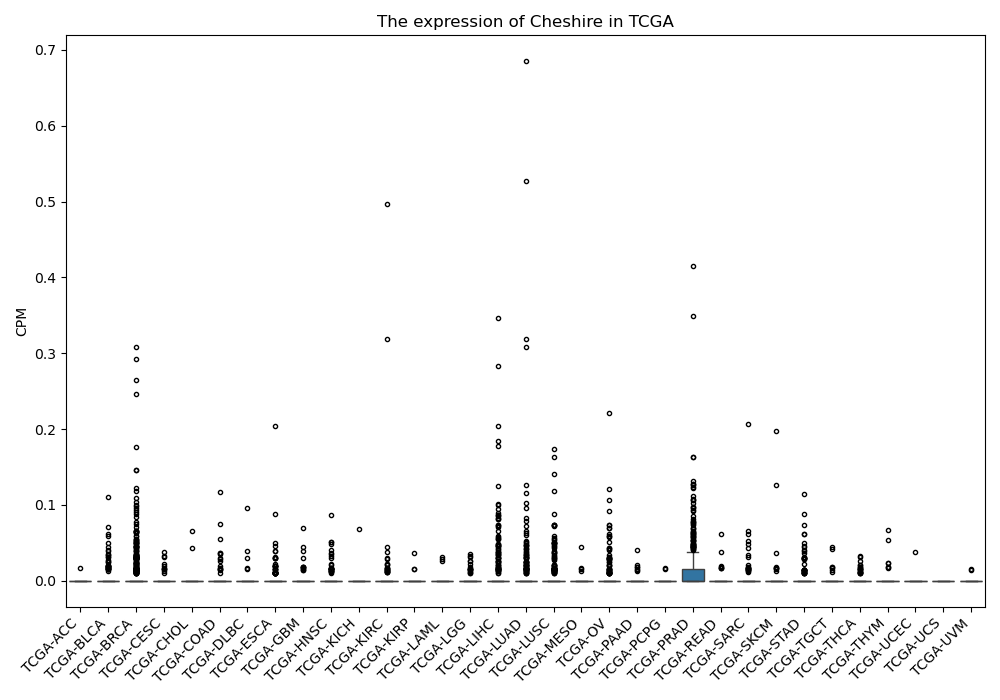
Cheshire
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000109 |
|---|---|
| TE superfamily | Charlie |
| TE class | DNA |
| Species | Eutheria |
| Length | 2420 |
| Kimura value | 18.40 |
| Tau index | 0.0000 |
| Description | hAT-Charlie DNA transposon, Chesire subfamily (autonomous) |
| Comment | 16bp TIR, 8bp TSD. |
| Sequence |
CAGGGGTCGGCAAACTACGGCCCGCGGGCCAAATCCGGCCCGCCGCCTGTTTTTGTACGGCCCGCGAGCTAAGAATGGTTTTTACATTTTTAAATGGTTGAAAAAAAAATCAAAAGAAGAATAATATTTCGTGACACGTGAAAATTATATGAAATTCAAATTTCAGTGTCCATAAATAAAGTTTTATTGGAACACAGCCACGCTCATTCGTTTACGTATTGTCTATGGCTGCTTTCGCGCTACAACGGCAGAGTTGAGTAGTTGCGACAGAGACCGTATGTGGCGCTCTCATCGCTTCGCACTGCTGCTCGGCGCACTACAAATCGCAGTGACGCAGTTATAACTCGACAGCGTTTCGAGTGCCACGCGTATCGCCGTACCGCGACATTTTTTTTTATTACCAGTGCATACCCATCATGTCAAAACAAGAAAAGAAGAGAAAAGTGGACTTCGAATGTCGCGCTTTTAAGGCACAGTGGAGTGTGGATTATTTTGTTATCGAATTAGATGGCAAAGCATTGTGTTTATTATGCAATGACACTATAGCTGTGCTAAAAGAATACAATATACGTCGACATTACCAGACTAAGCACTCATCACAATATTCCCAACTCACAGGAAAGCAACGGTCAGAAAAATTAGAAAATTTAAAACGGAATATCTCATCACAGCAGAATTTCTTCACAAAAATAAAAAATGAAAATGAGGCTGCAACCAAAGTAAGTTTCCGAGTGGCTCATTTGTTAGCCAAGCAAGGAAAGCCGTTTACCGATGGTGAGTTAATTAAATCGTGTTTGATTGCAGCAGCCGAAGAAATGTGTCCAGAGAAAATAAACTTGTTTAAGACTATTAGCCTTTCGGCGAGAACAGTTGCTCGAAGAGTTGAGGACATTGGGAGCAACATCAATAGTCAATTAAAAAACAAGGCAAATGATTTCGAGTGGTTTTCCTTGGCTCTTGATGAGTCGACAGATGTTACCGATACTGCTCAGTTGTTTATTCGAGGAGTCAATGCCGAGTTTGAAGTGACTGAAGAATTAGCCTCTATGAATAGTCTGCGTGGAACAACTACAGGCGAGAATATTTTCAAAGAAGTTGAGAAAACACTAATTCAGTACAACCTGAAGTGGAATCTGCTAAGATGTGTTACAACTGATGGTGGTAAAAATATGTGTGGAGCAGAAAAAGGCTTAGTTGGACAAATTTACAAAGCTTGTGAAAATGTAAGGTGTTTAAAGCCTATGGTTATTCATTGTATTATTCATCAGCAGGTACTTTGCGGAAAATATTTGAATCTATCATGTGTTATTGAACCAGTAGTGTCAACGGTGAACTTCATTCGCTCTCGTGGACTTAACCATCGTCAGTTCCGTGAATTTTTGTCAGAAATAGAAGCTGAATATCCTGACTTGCCCTACCACACAGCAGTTCGATGGCTTAGCAGTGGTAAAGTTTTATTGCGATTTTTTGAGCTCAGGGCCGAGATTGAAATTTTTCTGAATGAGAAGAACCGCCCTCAACCACTATTATCGAACACTGAATGGCTTTGGAAATTAGCTTTTGCTGCAGACTTGATAATGTTTCTTAATGAATTCAACCTAAAATTACAAGGCAAAACAGCGCTTATATGCGAAACTTATACTGCGGTAAAGTCATTTCGATGACAACAACTATTGTTTGAATCACAAGTAATGTCAAGCTGCTTTATACACTTCCCGTGCTGTCAAAAGTTAAAACAAGAAGCGAGATCTCCATTCCCACACAAATTTGCAGCGGATATATTTTCCGAGCTCAAACTACAGTTCCAGCAGCGTTTTTCGGACCTCGATGCAAGTGCAAAGGAAATTTCCATATTTCAAAATCCATTTAACTGTGCAATTGAGGAGCTTCCACCTAACCTTCAATTGGAAGTGATTAATCTGCAATGTAATGACATGCTAAAAGGCAAATATCAAGAGAAGAATCTAATAGAATTCTATAAATGCCTTCCAAGCGATGAATATGCTCAATTAAAATCATATGCTCGTGGATTGATATCAGTATTTGGCAGTACCTATCTGTGTGAAAAGACATTTTCAAAGATGAAATACGTAAAATCTCATTACAGATCAGCATTAACAGATGAACATTTGCAATCGATTTTGATGATAGGGAACACTAACTTTGAACCCCAATTAAGCGAAATGTTATCCCCCCAAAAAGAATTCCATTCTTCTCATTAGTAGACCTGTATTACAAAAAATTGTACTCAATTATTATTATTATTATATTTTGAATTTCGTCAATAAAAATTTTGTGGAAATTTGTTTTCTCTCTTGTTATATAAGTACCTACATAATATCCTCGATTTTGCCTCTTGGCCCGCAAAGCCTAAAATATTTACTATCTGGCCCTTTACAGAAAAAGTTTGCCGACCCCTG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Cheshire | CDF5 | 100 | 120 | - | 16.35 | TCTTCTTTTGATTTTTTTTTC |
| Cheshire | TSO1 | 146 | 160 | + | 15.97 | TATATGAAATTCAAA |
| Cheshire | TCX3 | 2268 | 2279 | - | 15.90 | AAATTCAAAATA |
| Cheshire | CDX1 | 393 | 402 | - | 15.81 | GGTAATAAAA |
| Cheshire | lin-54 | 2269 | 2281 | - | 15.79 | CGAAATTCAAAAT |
| Cheshire | MYB3R4 | 624 | 635 | - | 15.62 | TTCTGACCGTTG |
| Cheshire | HOXB13 | 182 | 190 | - | 15.62 | CCAATAAAA |
| Cheshire | ERF118 | 40 | 47 | + | 15.56 | CCGCCGCC |
| Cheshire | DOF5.8 | 1725 | 1743 | - | 15.42 | CTTCTTGTTTTAACTTTTG |
| Cheshire | NAC031 | 1674 | 1692 | + | 15.42 | TTGTTTGAATCACAAGTAA |
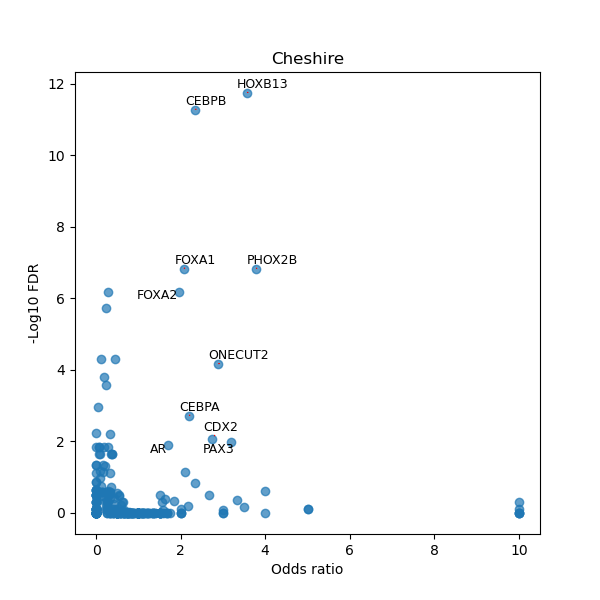
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.