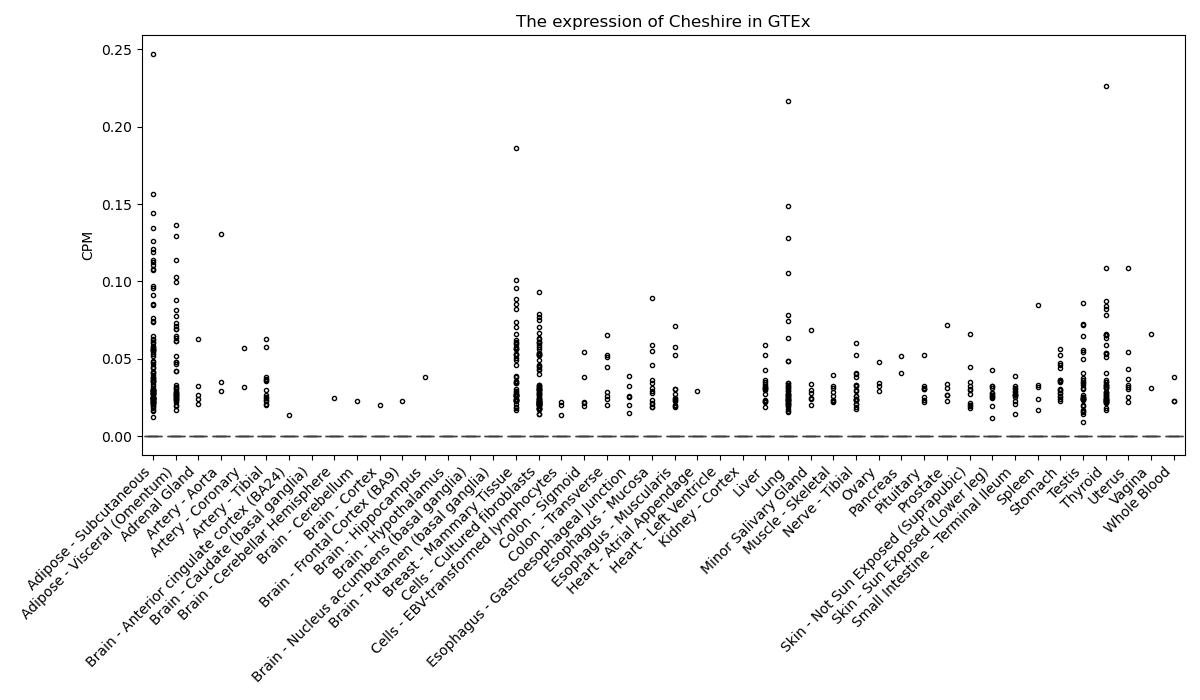
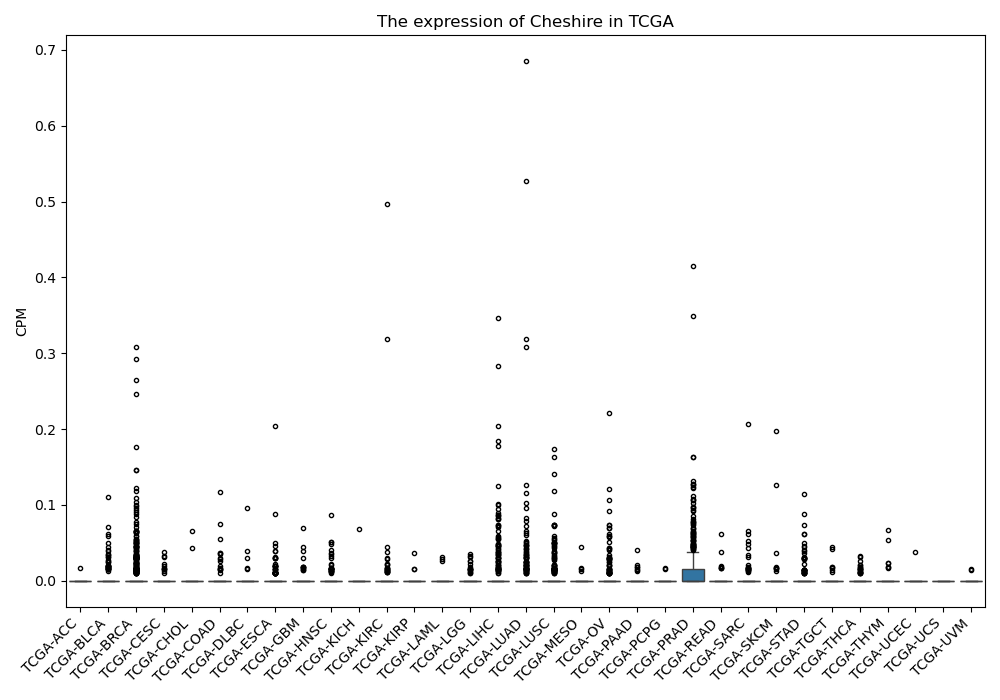
Cheshire
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000109 |
|---|---|
| TE superfamily | Charlie |
| TE class | DNA |
| Species | Eutheria |
| Length | 2420 |
| Kimura value | 18.40 |
| Tau index | 0.0000 |
| Description | hAT-Charlie DNA transposon, Chesire subfamily (autonomous) |
| Comment | 16bp TIR, 8bp TSD. |
| Sequence |
CAGGGGTCGGCAAACTACGGCCCGCGGGCCAAATCCGGCCCGCCGCCTGTTTTTGTACGGCCCGCGAGCTAAGAATGGTTTTTACATTTTTAAATGGTTGAAAAAAAAATCAAAAGAAGAATAATATTTCGTGACACGTGAAAATTATATGAAATTCAAATTTCAGTGTCCATAAATAAAGTTTTATTGGAACACAGCCACGCTCATTCGTTTACGTATTGTCTATGGCTGCTTTCGCGCTACAACGGCAGAGTTGAGTAGTTGCGACAGAGACCGTATGTGGCGCTCTCATCGCTTCGCACTGCTGCTCGGCGCACTACAAATCGCAGTGACGCAGTTATAACTCGACAGCGTTTCGAGTGCCACGCGTATCGCCGTACCGCGACATTTTTTTTTATTACCAGTGCATACCCATCATGTCAAAACAAGAAAAGAAGAGAAAAGTGGACTTCGAATGTCGCGCTTTTAAGGCACAGTGGAGTGTGGATTATTTTGTTATCGAATTAGATGGCAAAGCATTGTGTTTATTATGCAATGACACTATAGCTGTGCTAAAAGAATACAATATACGTCGACATTACCAGACTAAGCACTCATCACAATATTCCCAACTCACAGGAAAGCAACGGTCAGAAAAATTAGAAAATTTAAAACGGAATATCTCATCACAGCAGAATTTCTTCACAAAAATAAAAAATGAAAATGAGGCTGCAACCAAAGTAAGTTTCCGAGTGGCTCATTTGTTAGCCAAGCAAGGAAAGCCGTTTACCGATGGTGAGTTAATTAAATCGTGTTTGATTGCAGCAGCCGAAGAAATGTGTCCAGAGAAAATAAACTTGTTTAAGACTATTAGCCTTTCGGCGAGAACAGTTGCTCGAAGAGTTGAGGACATTGGGAGCAACATCAATAGTCAATTAAAAAACAAGGCAAATGATTTCGAGTGGTTTTCCTTGGCTCTTGATGAGTCGACAGATGTTACCGATACTGCTCAGTTGTTTATTCGAGGAGTCAATGCCGAGTTTGAAGTGACTGAAGAATTAGCCTCTATGAATAGTCTGCGTGGAACAACTACAGGCGAGAATATTTTCAAAGAAGTTGAGAAAACACTAATTCAGTACAACCTGAAGTGGAATCTGCTAAGATGTGTTACAACTGATGGTGGTAAAAATATGTGTGGAGCAGAAAAAGGCTTAGTTGGACAAATTTACAAAGCTTGTGAAAATGTAAGGTGTTTAAAGCCTATGGTTATTCATTGTATTATTCATCAGCAGGTACTTTGCGGAAAATATTTGAATCTATCATGTGTTATTGAACCAGTAGTGTCAACGGTGAACTTCATTCGCTCTCGTGGACTTAACCATCGTCAGTTCCGTGAATTTTTGTCAGAAATAGAAGCTGAATATCCTGACTTGCCCTACCACACAGCAGTTCGATGGCTTAGCAGTGGTAAAGTTTTATTGCGATTTTTTGAGCTCAGGGCCGAGATTGAAATTTTTCTGAATGAGAAGAACCGCCCTCAACCACTATTATCGAACACTGAATGGCTTTGGAAATTAGCTTTTGCTGCAGACTTGATAATGTTTCTTAATGAATTCAACCTAAAATTACAAGGCAAAACAGCGCTTATATGCGAAACTTATACTGCGGTAAAGTCATTTCGATGACAACAACTATTGTTTGAATCACAAGTAATGTCAAGCTGCTTTATACACTTCCCGTGCTGTCAAAAGTTAAAACAAGAAGCGAGATCTCCATTCCCACACAAATTTGCAGCGGATATATTTTCCGAGCTCAAACTACAGTTCCAGCAGCGTTTTTCGGACCTCGATGCAAGTGCAAAGGAAATTTCCATATTTCAAAATCCATTTAACTGTGCAATTGAGGAGCTTCCACCTAACCTTCAATTGGAAGTGATTAATCTGCAATGTAATGACATGCTAAAAGGCAAATATCAAGAGAAGAATCTAATAGAATTCTATAAATGCCTTCCAAGCGATGAATATGCTCAATTAAAATCATATGCTCGTGGATTGATATCAGTATTTGGCAGTACCTATCTGTGTGAAAAGACATTTTCAAAGATGAAATACGTAAAATCTCATTACAGATCAGCATTAACAGATGAACATTTGCAATCGATTTTGATGATAGGGAACACTAACTTTGAACCCCAATTAAGCGAAATGTTATCCCCCCAAAAAGAATTCCATTCTTCTCATTAGTAGACCTGTATTACAAAAAATTGTACTCAATTATTATTATTATTATATTTTGAATTTCGTCAATAAAAATTTTGTGGAAATTTGTTTTCTCTCTTGTTATATAAGTACCTACATAATATCCTCGATTTTGCCTCTTGGCCCGCAAAGCCTAAAATATTTACTATCTGGCCCTTTACAGAAAAAGTTTGCCGACCCCTG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Cheshire | E2F7 | 232 | 245 | + | -2.90 | CTTTCGCGCTACAA |
| Cheshire | NR1H2::RXRA | 1318 | 1334 | + | -3.27 | TAGTGTCAACGGTGAAC |
| Cheshire | THI2 | 105 | 119 | + | -5.62 | AAAAATCAAAAGAAG |
| Cheshire | Nr2F6 | 2158 | 2172 | - | -6.37 | GGGGTTCAAAGTTAG |
| Cheshire | Sox21b | 2200 | 2214 | + | -6.80 | AAGAATTCCATTCTT |
| Cheshire | DAL81 | 32 | 50 | - | -13.16 | ACAGGCGGCGGGCCGGATT |
| Cheshire | BCL6B | 1341 | 1357 | + | -14.60 | CGCTCTCGTGGACTTAA |
| Cheshire | BCL6B | 996 | 1012 | + | -17.40 | TTTATTCGAGGAGTCAA |
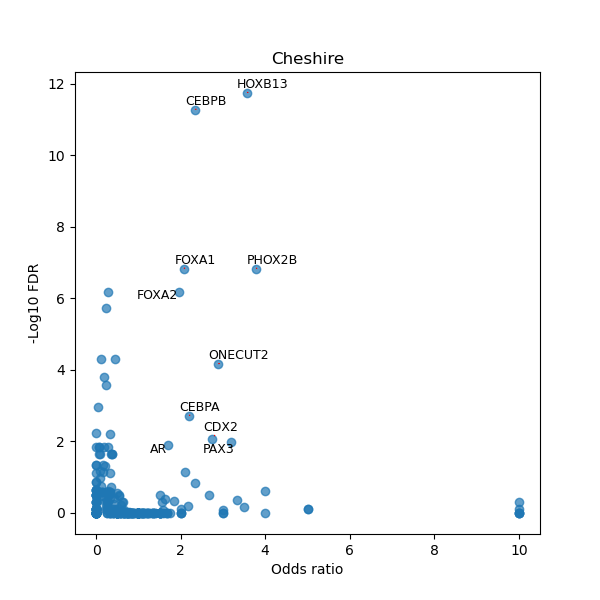
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.