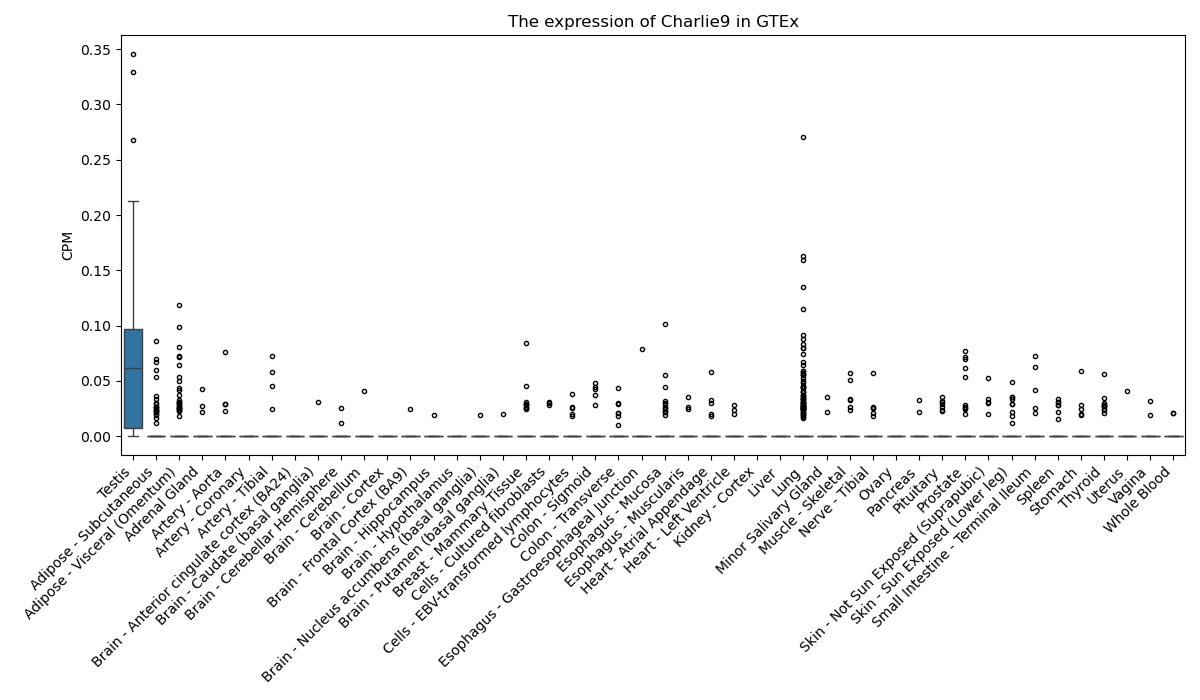
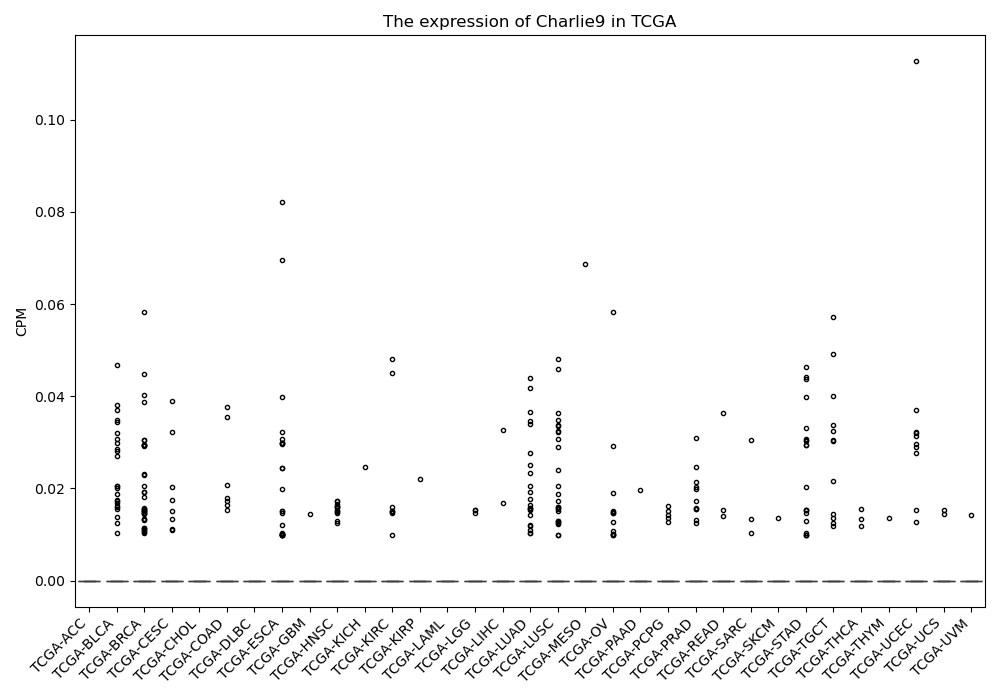
Charlie9
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000108 |
|---|---|
| TE superfamily | Charlie |
| TE class | DNA |
| Species | Eutheria |
| Length | 2787 |
| Kimura value | 23.76 |
| Tau index | 1.0000 |
| Description | hAT-Charlie DNA transposon, Charlie9 subfamily (autonomous) |
| Comment | Present in all eutherians, but absent in opossum & monotremes |
| Sequence |
CAGTGTTTTTCAAACTGCGGGTCGCGACCCATTAGTGGGTCGTGAAATCAATTTAGTGGGTCGCAACCAGCATTTTTAAAAGTAAAATGAAATAGAATAGAATAGAAAATATCAGAGTGCATCGCACGGATCCTTAGCAAGAACTGCGCACATTGCAACAAATACCGCGGCCAATTGCAATGNGCGAGAAGACAGTTAGTGGAGGGGATGTGATGAANACACGCATATTGGCTAGGAATGGGTCATTCGATCTTTTGCTCACTGGCATCTCNTTNGATTTTCNGGTATACGATGGTCAATTTGATCATCACCGTAACGGGAGGGTTAACGTTGTACAGTTTAATTTTATATTAACATAATTTGCAGTTAGTTGTGAAGCGAAGTGCGGGAGAATTTTATCCGCTGTCGGCAAAANTGTCGAGACTTTCAACCGAGTTTCGGTTACGAACTTTCAGTGTGTTTTTCAGAAAGGACGTTATCGAGTGTGACATTTTTCAAGTTCGTGACAATTGTAACATTTTAATCGAACTAGTACAAATTGCCCTTAAAATGGATCGCTTTATTATTAGAAGAAAAAAGAATGATGAAGTGAAAAATACAGAGGCATGTTCAAGTTCAACTGCTGGATCTGGAAGTGTAAATAGTGACAATATTGAGAAAAATGTTGACTCTAATCAGCAAACTTCGACTTCATTTGAGCCGCATTCTAAAAAGAAAAAAGTAAGTGCAAGACTTTATAATGAAGATTATTTAAAATATGGTTTTATCAAATGTGCAAAACCCANTGAAAATGACAGACCCCAATGTATTATTTGTAATGATATTCTTGCAAATGAAAGCTTAAAACCTTCAAAATTAAAAAGACACTTGGAAACACAGCATGCTGAACTTGTCGATAAGCCCCTCGAATATTTTCAAAGAAAGAAAAAAGACGTAAAGTTATCGGCACAATTTCTTAGTCGTTCTACGACTGTTAATGAGAAAGCCTTATTATCATCATATTTAGTTGCATATCGTGTGGCAAAAGAGAAAATGGCTCACACAGCTGCTGAAAAAATTATTCTTCCAGCATGTATGGATATGGTGCGTACGATTTTTGATGATAAATCAGCTGATAAATTAAAAACTATACCTCTTAGTGATAACACAGTATCTCGTCGAATCTNTACTATTGCAGAACATTTAGAAGCAATGCTTATTACGCGGTTACAGTCTGGTATAGATTTTGCGATCCAACTCGATGAGAGCACTGATATTGCAAGTTGCGCAACACTTTTAGTTTATGTCAGATATGCGTGGCAAGACGATTTTATGGAGGATTTGTTGTGTTGTTTAAATTTAACCTCACACACAACTGAATTAGACATATTTACAGAATTGGAAAAGCGCATTGTTGGTCAANATAAATTAAACTGGAAAAACTGTAAAGGAATNACGAGTGATGGAATAGCAAATATGACTGGAAAACACGGNAGAGNTATTAAAAAATTGTTAGAAGCTACTAATAACGGTGCTATTTGGAATCACTGTTTTATACATCGCGAAGCTTTGGCGTCCAAAGAAATTCTACCGAATCTCATGGATATATTGAAAAACGCAGCGAAAGTTGTTAATTTTATTAAAGGAAGCTCACTGAATAGCCGACTTTTTGAAATATTTTGTTCAGAGATTGGAGCTAACCATNCCCGCTTACTGTATCANACCGAAGTTCGTTGGTTGTCGCGAGGGAAAGTGCTAAGCAGGGTGTATGAACTCAGGAATGAGATTCACATTTTTCTAATTGAAAAGCAATCTCATTTGGCAAATATTTTCGAAGATGATATTTGAGTAANAAAATTGGCATATTTAACTGATATTTTCGGCATTCTTAACGAACTAAATTTGAAACTNCAGGGGAAAAACAACGATATATTTCAACATGTCGAACATATCCAAGGATTCCAAAAGACATTATTGTTATGGCAAGCAAGACTTAAAAGTAATCGCCCTAGCTACTATATGTTCCCAACATTGTTGCAACATATCGAAGAGAACATTATTAATGAAGACAGTTTGAANGAAATAAAATTAGAGATATTGTTGCATCTCACTTCTCTGTCTCAAACTTTTAATCATTACTTTCCAGAAGAGAAATTTGAAACATTAAAGAAAAATANTCGGATAAAAGATCCATTTGCTTTTCAAAACCCTGAATCAATAATTGAGTTAAACTTGGTGCCCGAAGAAGAGAATGAATTATTGCAGCTTAGTTCTTCATTTACACTGAAGAATGATTATAAGACATTAAGTNTATCATTATTTTGGATTAAGATTAAAGAAGAATTTCCATTGCTAAGTAGAAAGAGTGTATTGCTATTATTACCATTCACAACGACCNATTTGTNTGAACTAGGATTTTCAACCTTGACCCAATTAAAAACAAAGGAAAGAAACAGGCTCAACAGTGCACCAGATATGCGNGTAGCATTGTCCTCGTGTGTTCCGGACTGGAATGAACTTATGAACAGGCAAGCACACCCGTCGCATTAAATAAAATCTTTCCTAATTTTTGTTTTTGTATTGTTTATTTTGGANTATTTTTCTATGTTGTATTTAAATATTAATAAAATCATATTTGATATTAACTTTATGTTTTTACTTTTATCATACGTAGTAAGAGTAAGTATTGTTTCATGAAACTTTGTTTCAGTTGATATNTGTGCGTTATACGCGTATGTGTGTACTGGGTCGCGATGTAAAATGTATTTCTTACTGTGGGTCGCGGTCAAAAAAGTTTGAAAAACACTG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Charlie9 | crp | 1106 | 1117 | + | 16.89 | AATCAGCTGATA |
| Charlie9 | SOL1 | 2591 | 2605 | - | 16.85 | TATTAATATTTAAAT |
| Charlie9 | TCX6 | 2591 | 2605 | + | 16.85 | ATTTAAATATTAATA |
| Charlie9 | IRF6 | 434 | 442 | - | 16.60 | ACCGAAACT |
| Charlie9 | FOXE1 | 2560 | 2571 | - | 16.52 | CAAAATAAACAA |
| Charlie9 | DOF5.8 | 2625 | 2643 | + | 16.50 | CTTTATGTTTTTACTTTTA |
| Charlie9 | AP4 | 1107 | 1116 | + | 16.42 | ATCAGCTGAT |
| Charlie9 | AP4 | 1107 | 1116 | - | 16.42 | ATCAGCTGAT |
| Charlie9 | ATHB-40 | 2193 | 2203 | + | 16.20 | ATCAATAATTG |
| Charlie9 | HAT5 | 2195 | 2203 | + | 16.17 | CAATAATTG |
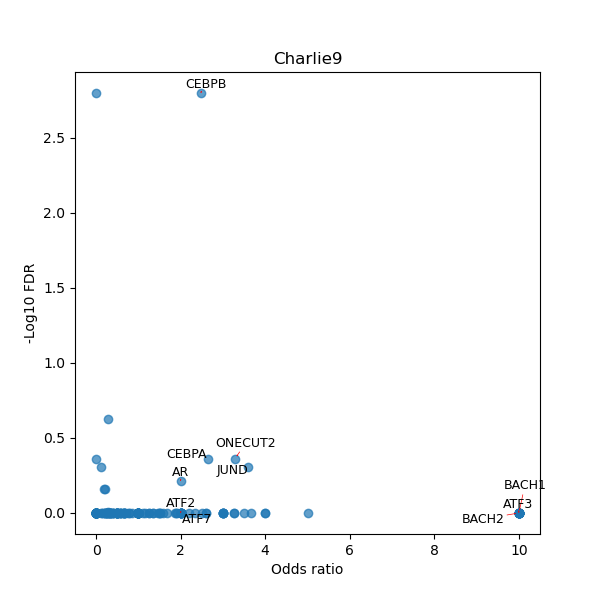
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.