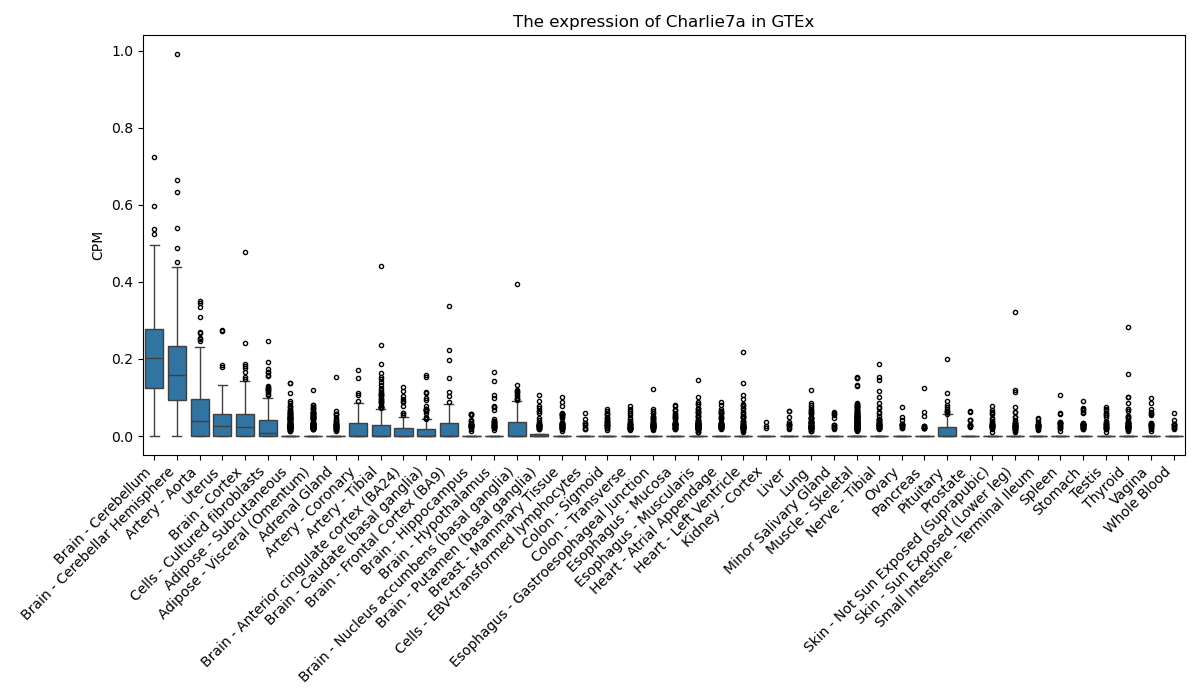
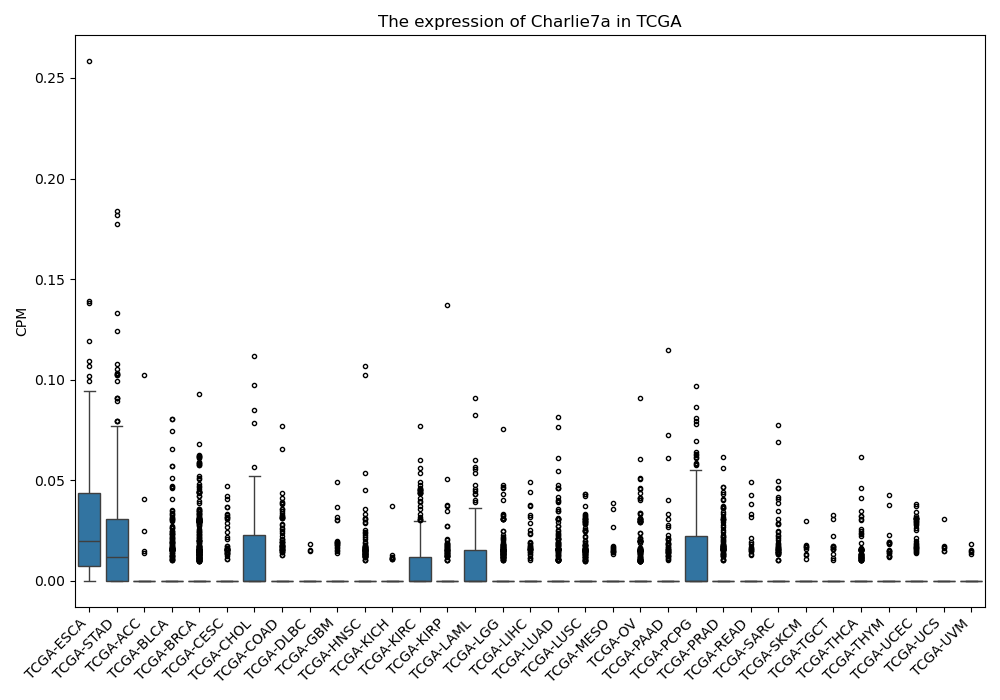
Charlie7a
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000106 |
|---|---|
| TE superfamily | Charlie |
| TE class | DNA |
| Species | Eutheria |
| Length | 272 |
| Kimura value | 27.68 |
| Tau index | 0.9720 |
| Description | hAT-Charlie DNA transposon, Charlie7a subfamily (non-autonomous) |
| Comment | Internal deletion product of Charlie7. |
| Sequence |
CAGTGGTCTCCAAAGTGGGGTGCGTGCACCCCAGGGGGTGTGCAAGACGATCCACTGGGGTGCGGGAAGAAAATATTAGAACTTCTATTTATATTTATTTTTATCCTTTAAAATTTCTATTTTTTGTGTATGTTTTATAATGTACATAATATATTAGTACAGTAGTACATGTATATAATTTATAAATAAATAAATATACATATATTGGGGGTGCATGCTCAAAAATTTTTTACTGATAGGGGTGCATGATCAAAAAAGTTTGGAGACCACTG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Charlie7a | sug | 29 | 40 | - | 12.92 | CACCCCCTGGGG |
| Charlie7a | ZBTB32 | 156 | 165 | + | 12.87 | AGTACAGTAG |
| Charlie7a | YER130C | 236 | 242 | - | 12.85 | CCCCTAT |
| Charlie7a | Cf2 | 197 | 205 | - | 12.81 | ATATATGTA |
| Charlie7a | Ebf4 | 28 | 38 | + | 12.81 | ACCCCAGGGGG |
| Charlie7a | FOXD3 | 187 | 200 | + | 12.79 | TAAATAAATATACA |
| Charlie7a | lsy-2 | 158 | 165 | - | 12.72 | CTACTGTA |
| Charlie7a | SPL8 | 136 | 151 | + | 12.63 | TATAATGTACATAATA |
| Charlie7a | FOXC2 | 183 | 193 | + | 12.51 | TAAATAAATAA |
| Charlie7a | TSO1 | 108 | 122 | - | 12.49 | AAATAGAAATTTTAA |
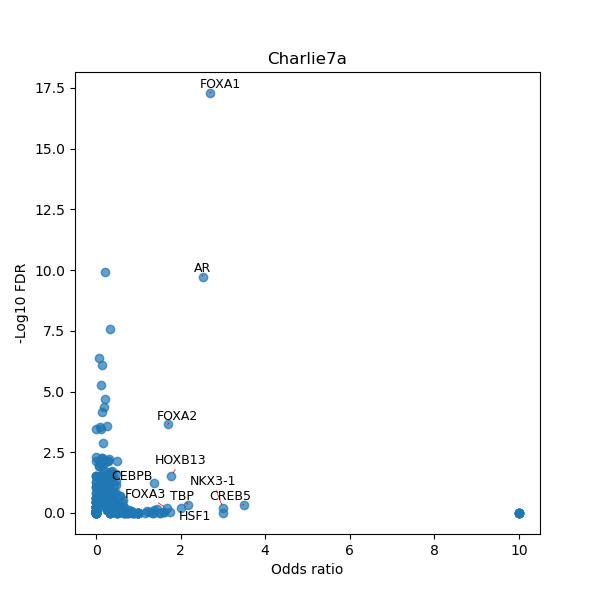
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.