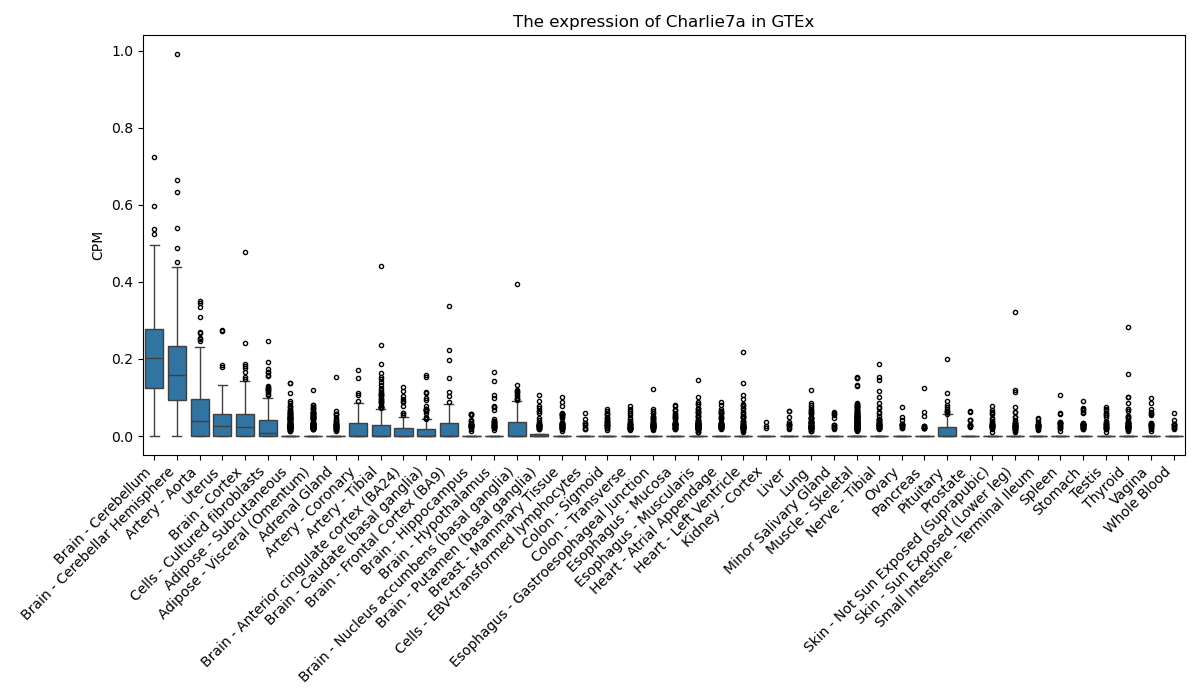
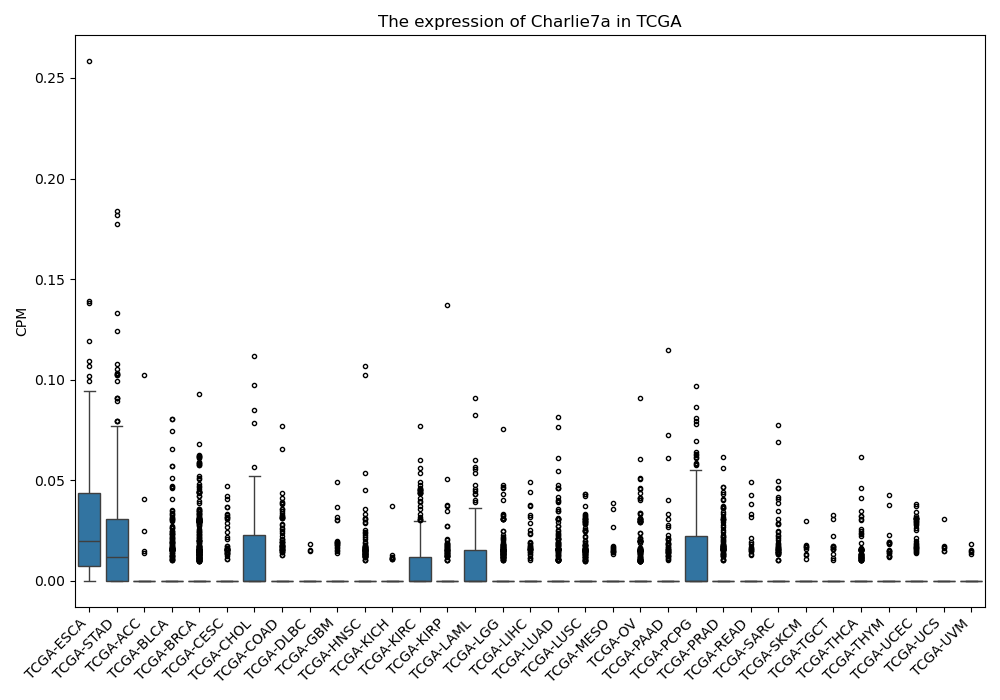
Charlie7a
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000106 |
|---|---|
| TE superfamily | Charlie |
| TE class | DNA |
| Species | Eutheria |
| Length | 272 |
| Kimura value | 27.68 |
| Tau index | 0.9720 |
| Description | hAT-Charlie DNA transposon, Charlie7a subfamily (non-autonomous) |
| Comment | Internal deletion product of Charlie7. |
| Sequence |
CAGTGGTCTCCAAAGTGGGGTGCGTGCACCCCAGGGGGTGTGCAAGACGATCCACTGGGGTGCGGGAAGAAAATATTAGAACTTCTATTTATATTTATTTTTATCCTTTAAAATTTCTATTTTTTGTGTATGTTTTATAATGTACATAATATATTAGTACAGTAGTACATGTATATAATTTATAAATAAATAAATATACATATATTGGGGGTGCATGCTCAAAAATTTTTTACTGATAGGGGTGCATGATCAAAAAAGTTTGGAGACCACTG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Charlie7a | ZIC1 | 27 | 40 | - | 13.51 | CACCCCCTGGGGTG |
| Charlie7a | AT3G10580 | 97 | 110 | - | 13.45 | TAAAGGATAAAAAT |
| Charlie7a | SOC1 | 114 | 126 | + | 13.37 | TTTCTATTTTTTG |
| Charlie7a | AT5G61620 | 97 | 110 | - | 13.32 | TAAAGGATAAAAAT |
| Charlie7a | Hmga1 | 97 | 104 | + | 13.22 | ATTTTTAT |
| Charlie7a | FLC | 113 | 126 | - | 13.16 | CAAAAAATAGAAAT |
| Charlie7a | TSO1 | 107 | 121 | - | 13.15 | AATAGAAATTTTAAA |
| Charlie7a | PI | 113 | 126 | - | 13.12 | CAAAAAATAGAAAT |
| Charlie7a | FOXC1 | 187 | 197 | + | 13.08 | TAAATAAATAT |
| Charlie7a | Crg-1 | 183 | 193 | + | 13.07 | TAAATAAATAA |
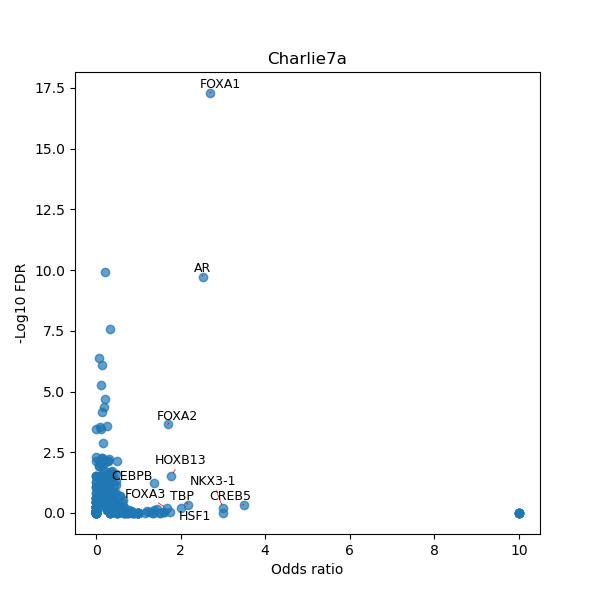
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.