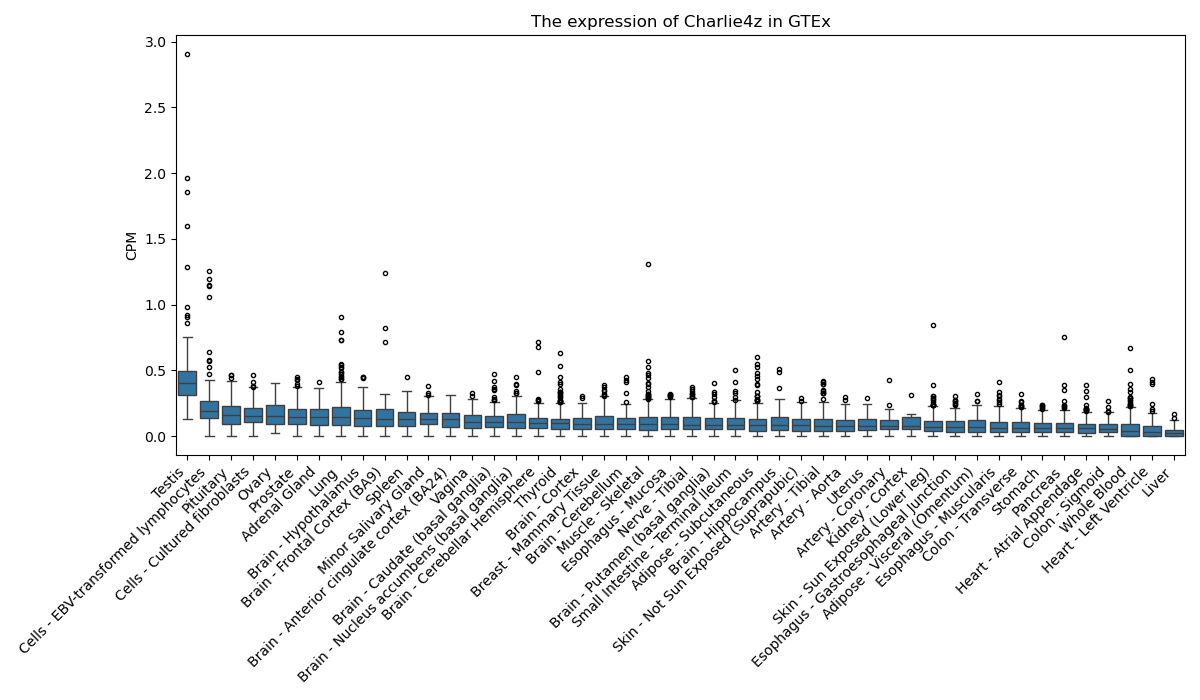
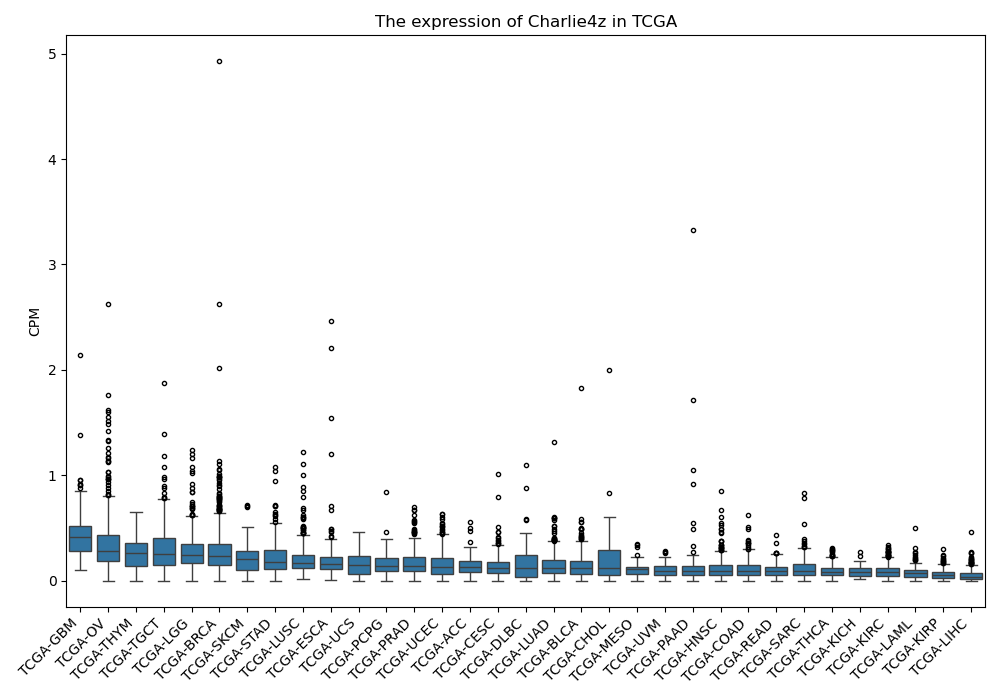
Charlie4z
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000026 |
|---|---|
| TE superfamily | Charlie |
| TE class | DNA |
| Species | Eutheria |
| Length | 167 |
| Kimura value | 29.90 |
| Tau index | 0.7613 |
| Description | hAT-Charlie DNA transposon, Charlie4z subfamily |
| Comment | 84% ID to MER80B. Not really an internal deletion product, but more like a "Ds" for the Charlie4 "Ac", having almost identical TIRs. |
| Sequence |
CAGGGGTTCTTAACCTGGGGTCCATGGATGGGCTTCAGGGGGTCCGTGAACCCCCTGAAATTGTATGCAAAATTTTGTGCGTATGTGCATTTTTCTGGGGAGAGGGTCCATAGCTTTCATCAGATTCTCAAAGGGGTCCGTGACCCCAAAAAGGTTAAGAACCACTG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Charlie4z | PAX5 | 138 | 145 | + | 12.09 | CCGTGACC |
| Charlie4z | POU2F1 | 64 | 72 | + | 12.05 | TATGCAAAA |
| Charlie4z | msn-1 | 35 | 41 | - | 12.03 | CCCCTGA |
| Charlie4z | msn-1 | 52 | 58 | + | 12.03 | CCCCTGA |
| Charlie4z | ZNF449 | 26 | 35 | - | 12.03 | AAGCCCATCC |
| Charlie4z | NR1H4::RXRA | 40 | 52 | - | 11.99 | GGTTCACGGACCC |
| Charlie4z | SFP1 | 69 | 78 | - | 11.97 | ACAAAATTTT |
| Charlie4z | RXRB | 40 | 53 | + | 11.95 | GGGTCCGTGAACCC |
| Charlie4z | POU5F1B | 64 | 72 | + | 11.85 | TATGCAAAA |
| Charlie4z | SOC1 | 145 | 157 | - | 11.65 | TAACCTTTTTGGG |
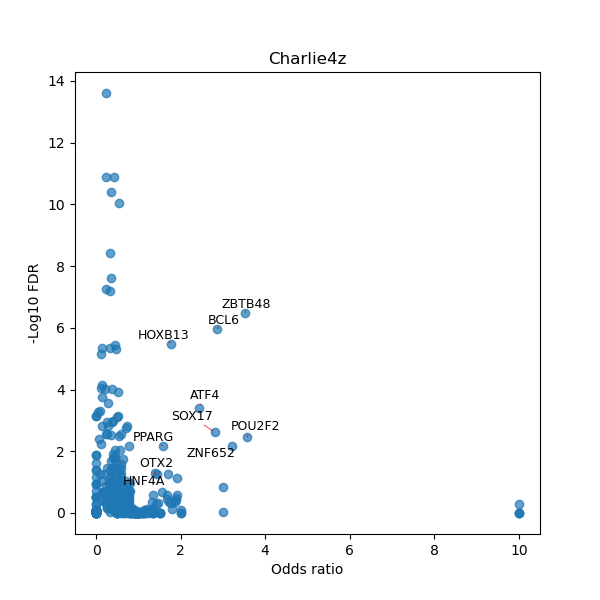
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.