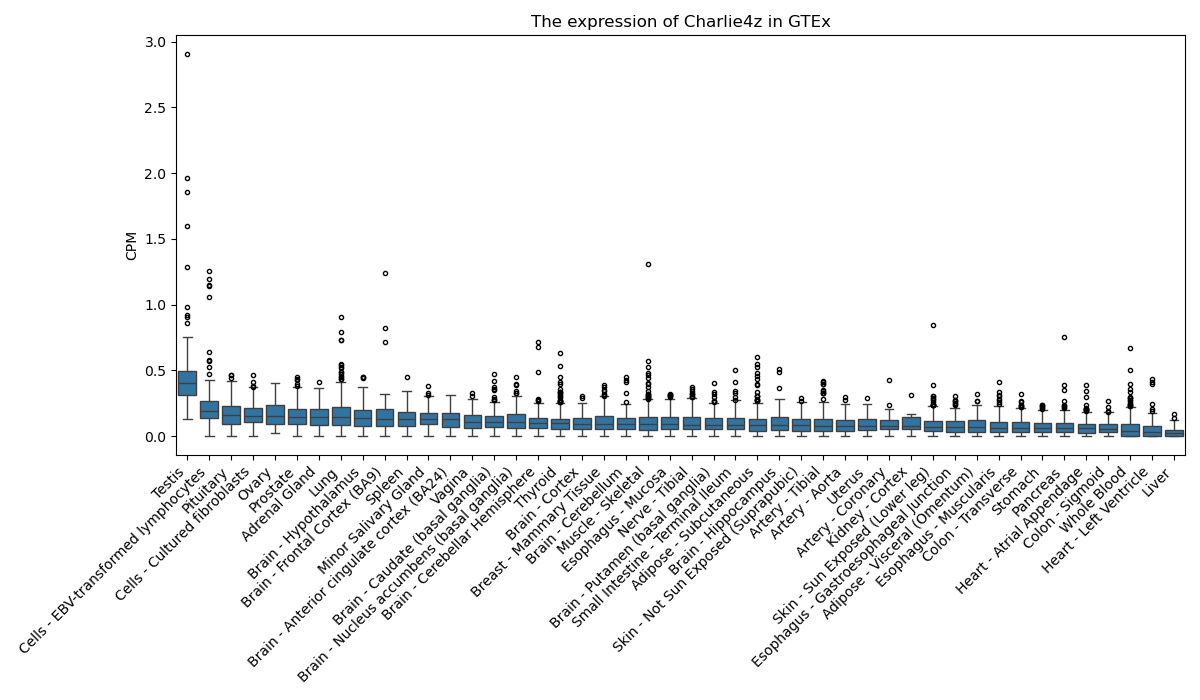
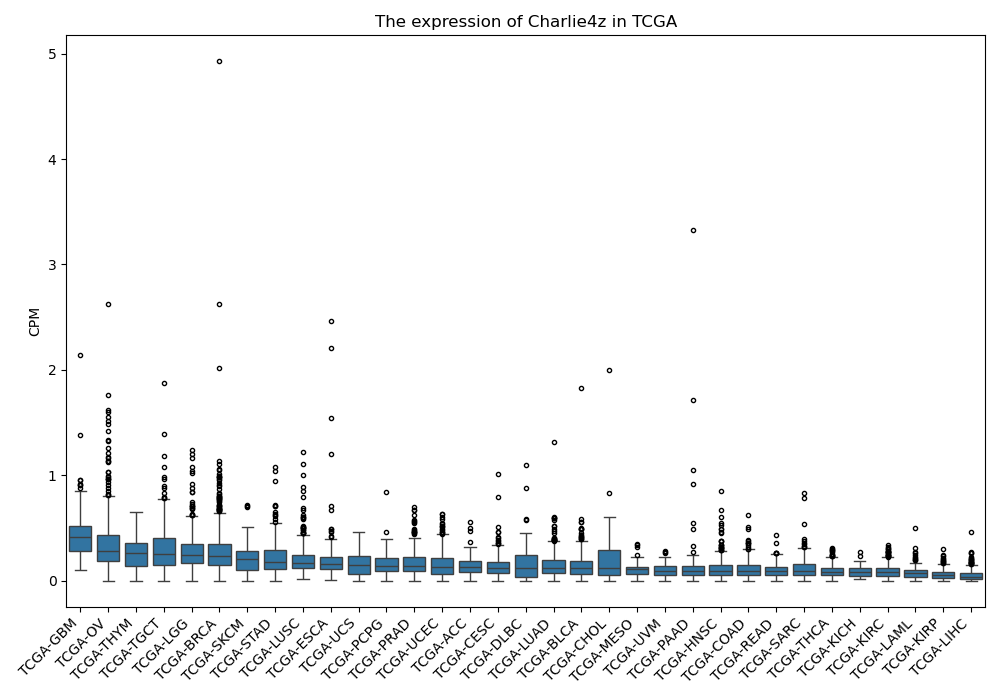
Charlie4z
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000026 |
|---|---|
| TE superfamily | Charlie |
| TE class | DNA |
| Species | Eutheria |
| Length | 167 |
| Kimura value | 29.90 |
| Tau index | 0.7613 |
| Description | hAT-Charlie DNA transposon, Charlie4z subfamily |
| Comment | 84% ID to MER80B. Not really an internal deletion product, but more like a "Ds" for the Charlie4 "Ac", having almost identical TIRs. |
| Sequence |
CAGGGGTTCTTAACCTGGGGTCCATGGATGGGCTTCAGGGGGTCCGTGAACCCCCTGAAATTGTATGCAAAATTTTGTGCGTATGTGCATTTTTCTGGGGAGAGGGTCCATAGCTTTCATCAGATTCTCAAAGGGGTCCGTGACCCCAAAAAGGTTAAGAACCACTG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Charlie4z | POU2F3 | 64 | 72 | + | 12.79 | TATGCAAAA |
| Charlie4z | HHO6 | 122 | 128 | + | 12.63 | AGATTCT |
| Charlie4z | sug | 33 | 44 | - | 12.56 | GACCCCCTGAAG |
| Charlie4z | INSM1 | 49 | 60 | - | 12.50 | TTTCAGGGGGTT |
| Charlie4z | TB1 | 37 | 45 | - | 12.46 | GGACCCCCT |
| Charlie4z | POU5F1 | 65 | 71 | + | 12.44 | ATGCAAA |
| Charlie4z | nub | 64 | 74 | + | 12.41 | TATGCAAAATT |
| Charlie4z | RARB | 134 | 146 | + | 12.33 | GGGTCCGTGACCC |
| Charlie4z | rib | 66 | 74 | + | 12.31 | TGCAAAATT |
| Charlie4z | Stat5a::Stat5b | 93 | 101 | - | 12.18 | TCCCCAGAA |
TFBS enrichment in GRCh38
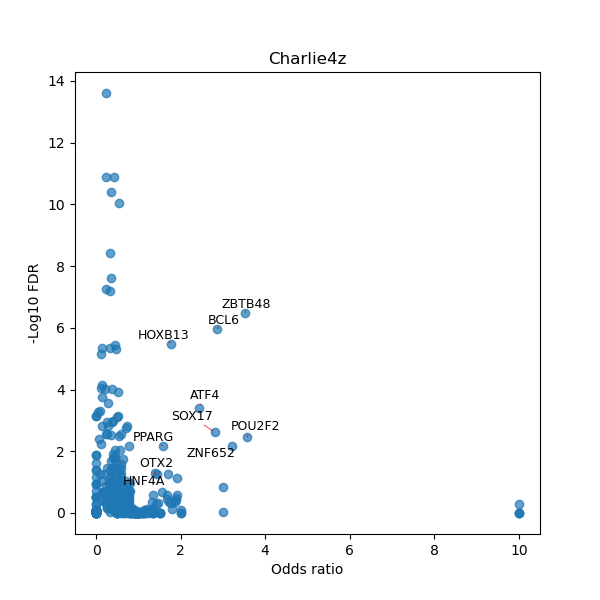
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.