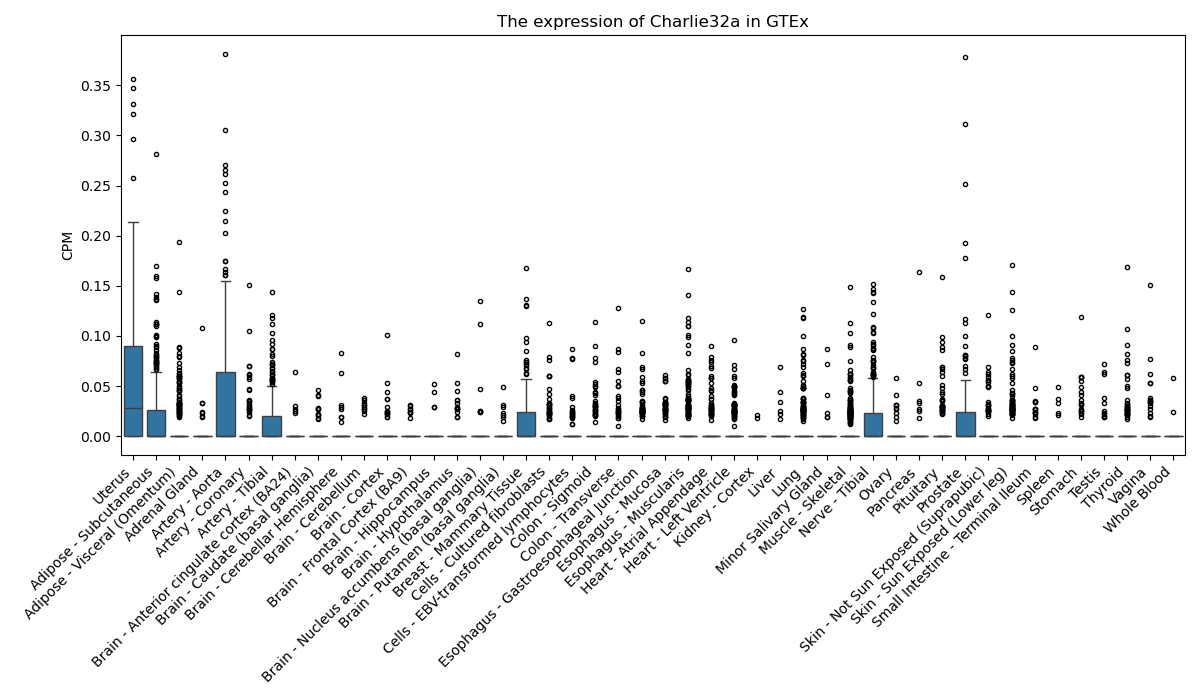
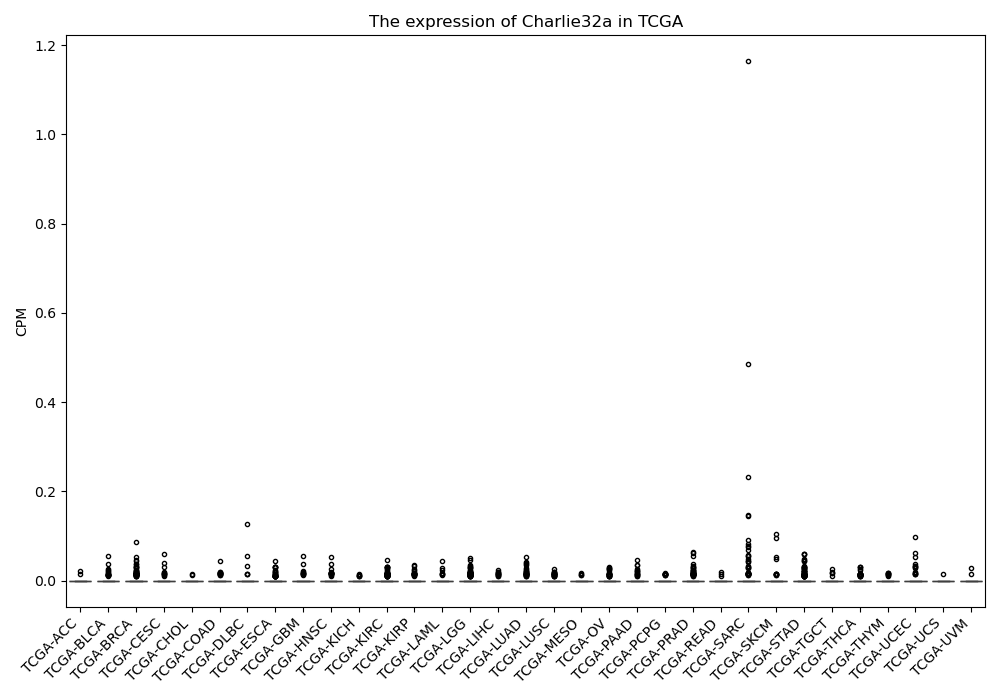
Charlie32a
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0001249 |
|---|---|
| TE superfamily | Charlie |
| TE class | DNA |
| Species | Theria_mammals |
| Length | 336 |
| Kimura value | 30.45 |
| Tau index | 1.0000 |
| Description | hAT-Charlie DNA transposon, Charlie32a subfamily |
| Comment | Present at orthologous sites in all therians, but absent from platypus. |
| Sequence |
CAGCGGTTCTCAAAGTGTGGTCCGGGGACCGCTGGTGGTCCGCGAGGGTCCTCCAGGTGGTCCGCGGGCTGGTCTCTTGTAGTATTAATGTTTATAATATGACAATGTCTATACTTTTGTTTCTGGCATGAATTAATAATTTACTTATGATATTTAATAAAAACCAGACATCACCTCATTGGTGTCTCATCAAAAGCAGCGCCATTCTTCCCATTAATTCTGGTGAGGTTTTTGTTCCTGTTGGTGAGGTGGCAGCGAGAATGGGCAGGATTATGGAGCTGGGGCTCTTTCCTCGGCTGAAGTGGTCCGCGGACCGAAAANGTTTGAGAACCACTG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Charlie32a | TCP4 | 57 | 64 | - | 6.40 | CGGACCAC |
| Charlie32a | TCP4 | 25 | 32 | + | 6.33 | GGGACCGC |
| Charlie32a | BZIP44 | 242 | 254 | + | 5.88 | TGGTGAGGTGGCA |
| Charlie32a | ZIC5 | 26 | 40 | - | 4.94 | GACCACCAGCGGTCC |
| Charlie32a | PDR3 | 61 | 68 | - | 3.75 | CCCGCGGA |
| Charlie32a | PDR3 | 61 | 68 | + | 3.75 | TCCGCGGG |
| Charlie32a | ZNF558 | 106 | 134 | - | 2.83 | AATTCATGCCAGAAACAAAAGTATAGACA |
| Charlie32a | GLI3 | 27 | 41 | - | 1.48 | GGACCACCAGCGGTC |
| Charlie32a | CG4360 | 114 | 126 | - | 0.69 | CCAGAAACAAAAG |
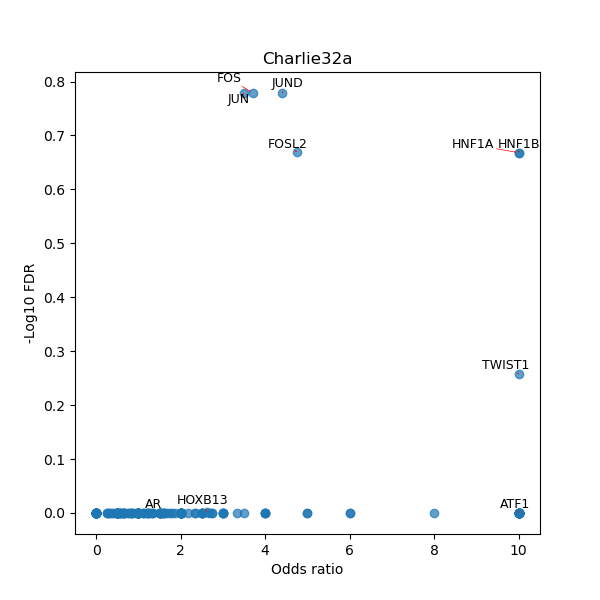
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.