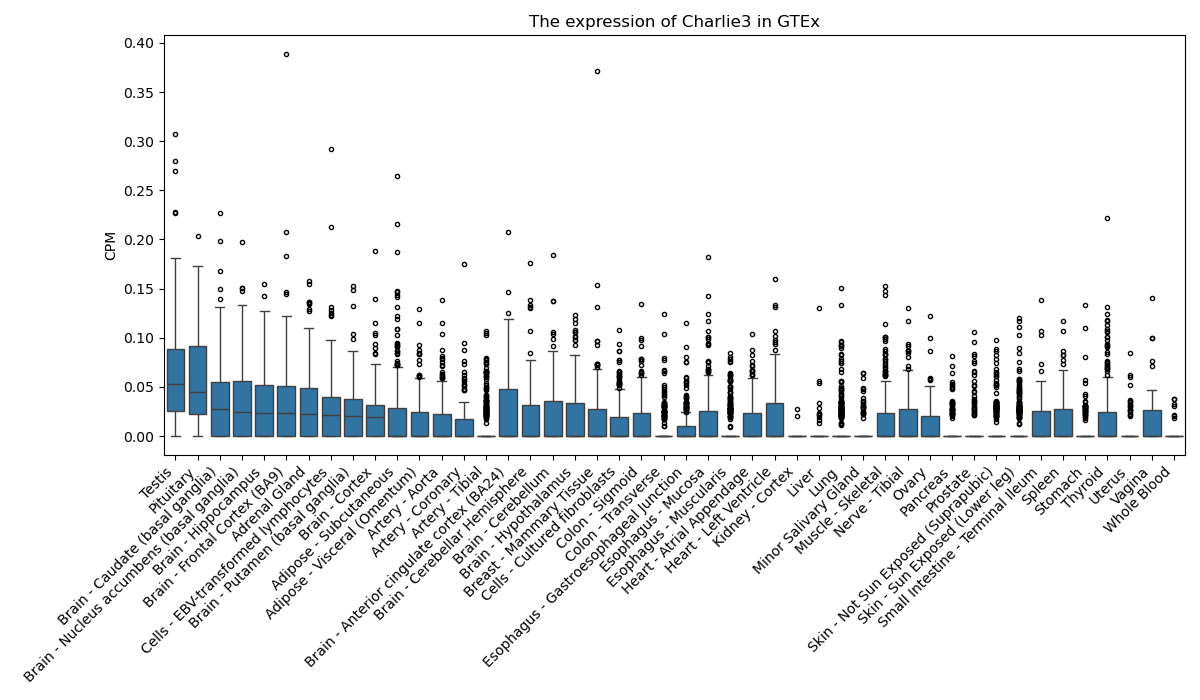
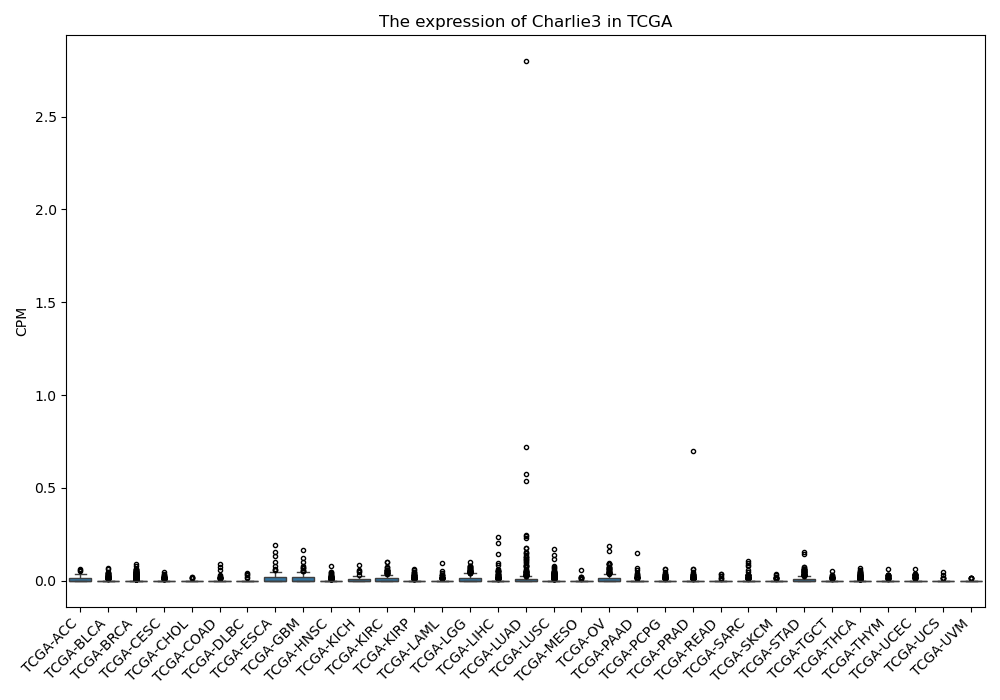
Charlie3
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000023 |
|---|---|
| TE superfamily | Charlie |
| TE class | DNA |
| Species | Simiiformes |
| Length | 2706 |
| Kimura value | 8.31 |
| Tau index | 0.9037 |
| Description | hAT-Charlie DNA transposon, Charlie3 subfamily |
| Comment | ORF (roughly 625-2526) encodes a transposase 59% identical to that of CHARLIE1. Common internal deletion products of CHARLIE3 are roughly: 1-272<>2456-2710 (MER1A), and 1-96<>135-272<>2621-2710 (MER1B). |
| Sequence |
CAGGGGTCCCCAACCCCCGGGCCACGGACCGGTACCGGTCCGTGGCCTGTTAGGAACCGGGCCGCACAGCAGGAGGTGAGCGGCGGGCGAGCGAGCGAAGCTTCATCTGTATTTACAGCCGCTCCCCATCGCTCGCATTACCGCCTGAGCTCCGCCTCCTGTCAGATCAGCGGCGGCATTAGATTCTCATAGGAGCGCGAACCCTATTGTGAACTGCGCATGCGAGGGATCTAGGTTGCGCGCTCCTTATGAGAATCTAATGCCTGATGATCTGAGGTGGAGCTGAGGCGGTGATGCTAGCGCTGGGGAGCGGCTGCAAATACAGATTAACATTAGCAGAGAGGTTTGACTGCACAGAGACCATAATAAATCAATTGCTTGCAGACTCATATCAAAACCCTATCAGTGAGTGGCAAGTGACAATTAAGCTGCATCTGGTGGCAGGCTTTATAGTGGCAAGTGAGTTGATGTACTTCAATTGTACAGCTGCATCTGGTGGCAGGCTTTAAGTCAGAATCCGACACTTATTTTAGTCCGCGCGTGGCCCGCCCATTATTTTATTTACCACTTCCGTCCGCGCCTCTTTCCCGCACTGCGCACTTGTCTCAGTCACAGTTTTGGTAAGCCCACAAGCTAACCCTAGCCAAAATGAGTAAAAAACAAACGTCACTGGAGAGCTTCTTTGAAAAGGGGGAAAGACCCAATGATGAGACAGCAGAAGACTCTAAGACTGCCAACAAAAAGAAAGCTGCATTTAAAAGAAAATACCAAGAGTCCTACTTAAATTACGGGTTCATTGCAACAGGTGATTCACATTCTCCAAGCCCGCTTTGTATAATATGTGGCGACCGGCTATCCAACGAAGCCATGAAACCTTCAAAACTGCTTCGCCACATGGAGACCAAGCACCCTGCATTAAAAGACAAGCCTTTGGAGTTTTTCAAAAGAAAAAAACGTGAACACGAAGAACAGAAGCAATTATTGAAGGCCACCACTTCATCAAATGTGTCTGCACTGAGAGCATCATTCTTAGTGGCTAACCGCATTGCTAAAGCTAAGAAGCCCTTTACTATTGGTGAAGAGTTGATCCTGCCTGCTGCTAAGGACATTTGTCGTGAACTTTTAGGAGAGGCTGCAGTTCAAAAGGTGGCACGTGTTCCTCTTTCGGCTAGCACCATAACTAGACGAATTGATGAAATAGCAGAGGATATTGAGGCACAATTGTTAGAGAGGATTAATGAGTCACCGTGGTACGCAATCCAGGTTGACGAGTCTACCGATGTTGACAACAAGGCAACAATGCTTGTTTTTGTGCGATATATTTTTCAGGAGGATGTGCATGAGGATATGTTATGTGCACTTTTGTTGCCAACCAACACCACAGCTGCAGAACTATTCAAGTCTTTGAATGATTACATATCAGGAAAACTGAATTGGTCATTTTGTGTCGGTATATGCACGGACGGAGCGGCTGCCATGACTGGACGGCTTTCTGGTTTCACTACTCGGGTCAAAGAGGTCGCTTCTGAATGTGAGTCTACGCACTGTGTCATCCATAGAGAAATGCTGGCTAGCCGAAAAATGTCACCTGAACTTAACAACGTTTTGCAGGATGTGATTAAAATTATCAACCACATTAAAGTACATGCCCTTAACTCACGTCTGTTCGCGCAGCTCTGTGAGGAGATGGACGCAGAGCACACACGTCTTCTCTTATACACAGAAGTGAGATGGCTTTCTAAAGGTAGATCACTGGCCAGAGTTTTTGAGTTACGAGAGCCGCTCCAGAGATTTCTTTTAGAAAAACAGTCACCACTGGCAGCACATTTCAGTGACACAGAATGGGTCGCAAAACTTGCTTACTTGTGTGACATATTCAACCTGCTCAACGAACTCAATCTGTCACTTCAGGGGAGAACGACAACTGTGTTCAAGTCGGCAGATAAAGTGGCTGCATTCAAAGCCAAACTGGAATTATGGGGGCGACGAGTGAACATTGGGATTTCTGACATGTTTCAAACATTAGCAGAGATTTTGAAAGAGACTGAGCCAGGGCCTTCTTTCTCCCAGCTGGTGCATGATCACCTATCTCAGCTTTCAAAAGAGTTTGAGCATTACTTCCCAACCACAAAAGACCCCCGAACTGGGAAGGAATGGATCCGCGACCCATTTGTGAATAAGCCAGGTGAATCGACTTTGTCCGTGCTAGAAGAGGATCAACTGCTTGAGATCGCAAATGACGGTGGCCTTAAAAGTATGTTTGAGACAACTTCAAATCTCCATACGTTCTGGATTAAAGTCAAGGCGGAATATCCTGAGATTGCCACAAAAGCACTGAAAAGCCTGCTTCCATTTCCAACATCCTATCTTTGTGAAGCAGGGTTTTCTGCAGTGACAGCAACCAAAACGAGATTACGGAGTAGACTGGACATAAGCAACACACTTCGGGTGTCACTGTCTCCCATCACCCCCAGATGGGACCGTCTAGTTGCAGGAAAACAAGCTCAGGGCTCCCACTGATTCTACATTATGGTGAGTTGTATAATTATTTCATTATATATTACAATGTAATAATAATAGAAATAAAGTGCACAATAAATGTAATGCGCTTGAATCATCCCGAAACCATCCCCCCCACCCCCGGTCCGTGGAAAAATTGTCTTCCACGAAACCGGTCCCTGGTGCCAAAAAGGTTGGGGACCGCTG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Charlie3 | RARA | 1866 | 1883 | - | -9.15 | AGGTTGAATATGTCACAC |
| Charlie3 | Sox21b | 396 | 410 | + | -9.89 | AACCCTATCAGTGAG |
| Charlie3 | EWSR1-FLI1 | 571 | 588 | - | -10.79 | GGAAAGAGGCGCGGACGG |
| Charlie3 | BCL6B | 217 | 233 | + | -16.61 | CGCATGCGAGGGATCTA |
| Charlie3 | DAL81 | 567 | 585 | - | -26.29 | AAGAGGCGCGGACGGAAGT |
| Charlie3 | BPC5 | 2028 | 2057 | + | -29.66 | AGAGATTTTGAAAGAGACTGAGCCAGGGCC |
| Charlie3 | BPC5 | 2026 | 2055 | + | -47.60 | GCAGAGATTTTGAAAGAGACTGAGCCAGGG |
| Charlie3 | DAL81 | 1086 | 1104 | - | -52.00 | CTTAGCAGCAGGCAGGATC |
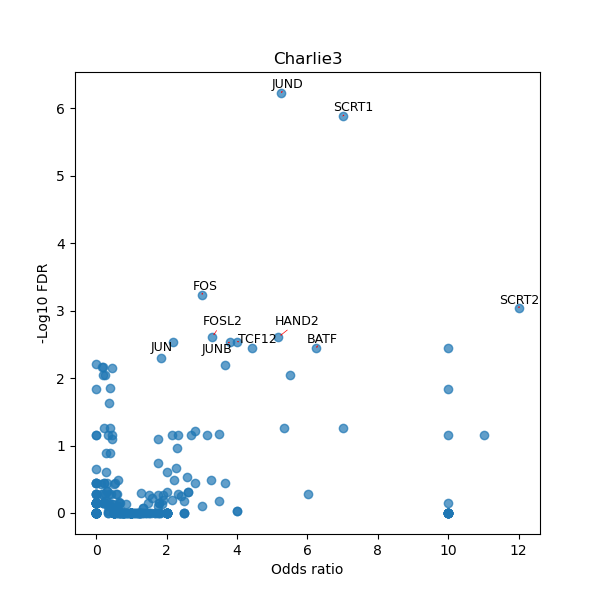
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.