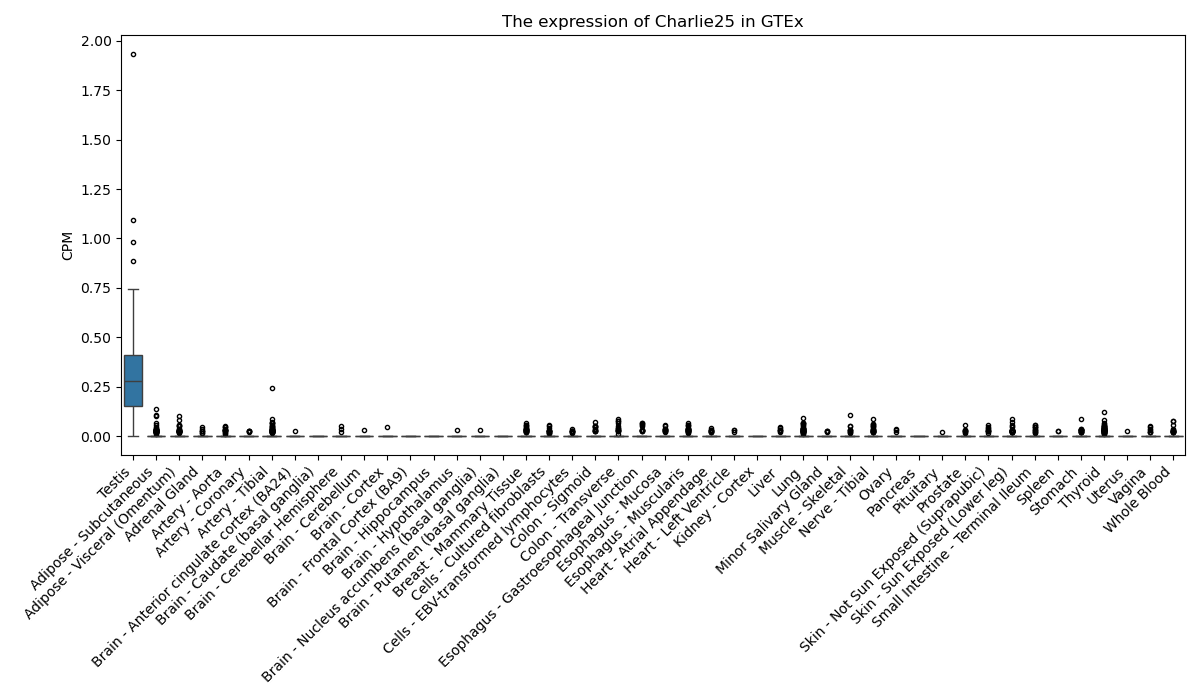
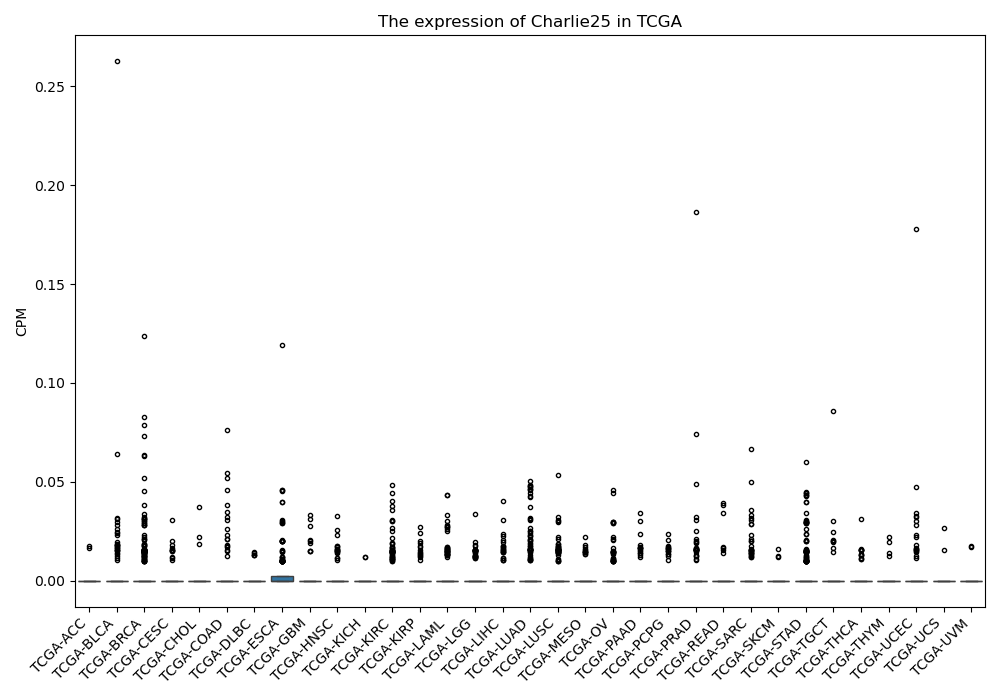
Charlie25
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000102 |
|---|---|
| TE superfamily | Charlie |
| TE class | DNA |
| Species | Eutheria |
| Length | 2524 |
| Kimura value | 30.06 |
| Tau index | 1.0000 |
| Description | hAT-Charlie DNA transposon, Charlie25 subfamily (autonomous) |
| Comment | 14 bp TIR. Complete ORF (roughly 384-2366) encodes a transposase closest to that of Charlie3 (44% identical, 62% similar). |
| Sequence |
CAGTGTTTCTCAAAGTGTGGTCCGCGGACCACTGGCGGTCCCCNGCGGTTCTATCAAGTGGTCCGCAGGCGGTTTGGCGGTTTCAGAGGAAAAAGCGATGAAACAATTTTGTTCACATACATTTCACAAATTTGAAATGTAAGATTAATTATGATTTTCACAGAAATCCCGTTACGTTCTTAATAATCGTTACGTTCTTAAAGGTTGCGCATGCGCTACAAGGACTGCGTTGGTCAGTTCGTCTCGGCTAACATTCAGTTAACAGGGTGCAGTTCGTCTCGGCTAACGTATTTTCACGTCATTTGCATGTTATTGTTTACGTTTGTTAAATTTGCATTTTTCGTTGTTACTATTGTGTTGTATTATATCCCCAATTCACAAAAATGGATCAATGGCTCAAAAGTGGTTCATTGAAGCGTAAAAGTAGTGATGAAAATAGTAACGTAAATACTACAACTCAGAATAACGTCATAAACGTAAATAGTGAACAGGACTCCAGTGCGAATATAGAATGTGAATCTGTATGTGCTGGGACAAGTGAATCTGCGAGTGTGATGATTTCGCACAAGCAGCCGAAAAAGAAAAGTGCGAATAGGAAGTACGACGATGAATATCTGAAAATTGGATTTTATTGGACCGGCGATCCATTTGCCCCTAGTCCCCAGTGCGTTGTCTGTTATGAAACTTTGTCAAATAGTGGCATGAAGCCATCGAAGCTTTCGCGTCATTTTCAAACAAAGCACAGTGACCTCTCTGGTAAACCAATCGAGTTTTTCCAGAACAAGCGCAAAATAATGCTTTCCAGTACGAAATTGATGAATTTTGTCGCTAAAGGCAGAGAAGAGACCAAAACTACAGAGGCATCATTCAAAGTTGCACTCCTTATAGCAAAAACAGGTACAAGTCACACTATTGCCGAGAAGCTTGTAAAACCAGCCGCAAAGTTAATGACAAATATTATGCTCGGAGAGAAAGCAGAACGAGCTATTGGCAAAATTCCTTTATCAAATGACACTGTTGGACGTCGCATAATATCAATGGCATACAATGTGGAAGAGCAATTACTATCACGTGTGCGTGCTAGCAGATATTTCGCTTTACAGTTGGACGAAACCACTGATGTGCAGAGTATGAATCAGCTCTTGGCATATGTGCGGTACATATATGAGGGAGAAGTGCTCGACGACTTTTTGTTCTGTTTATCACTGAAAACCCATGCTACAGGAGAAGATTTTTTTTATTTAGTTAACGATTATTTTGTGAGCCGTGATGTAGACTGGAAAAGGTGCGTTGGGATCAGTACTGACGGAGCACCAGCTATGTGTGCAGCAAGGAAGGGCGTTGCTACGCGAATAAAAGAGGTTGCACCTGAATGCCAATCCACACACTGCTTTATTCACAGAGAACAGTTGGCGGTCAACAATATGCCTCCTGATCTTGATTCAGTGTTGAAGGAAATAGTGAAAATTGTGAATACGATCAAATCGCGGCCACTGAGTGTACGTCTTTTCAGCGTGCTGTGCGAAGAAATGGGCAGCGAGTACAAGACTCTGCTTTTCCACACTGAAGTACGCTGGCTGTCGAGGGGAAAGGTGCTCACACGAGTTTTTGAAATGAGGGATGAGATAAAAACATTTCTTCATGACACTGATAATGCCAGTAAAGACCATTTCTATGATTTCAAGTGGCTTGCTCAAGTGGCGTATCTCAGCGATATATTCAGTATCTTGAATAGCCTGAACCTATCACTTCAGGGCCGAAATATCACGATTTTTAATGTTGAGGATAAGATATCAGGATTTCTTAAGAAGACCGAACTGTGGTGCAAACGGCTCGATCGCCGAGAGTTTGACTCTTTCCCAACACTTGATGATTTTCTTCACTCGTCGGAGAAAGAAATCGATGACGTATTATTGGGCATATTTAAAAACCACATCCAAATGCTGCAACAGAACATGAAGAAATACTTTCCAGAGCCGAATGCAACCAAAGAGTGGATTAGGAATCCATTCGCCGCTATCTCCCAAGTTGAAACATTCAACCTTCCAGCTTTTGAGTGTGATATGCTCGTCGACTTAGCGTCTGACGGAGCGCTGAAAGTAGTCTTCAGTGAGAAATCTCTCCATAATTTCTGGGTCCATGTTCGATCCGAATATCCAGAACTATCTGACAGAGCCACCAAACACTTGCTGCCATTCCCGACGACCTATAATTGTGAGTTAGGATTTTCTAATTTAGTAGAAATAAAGAGTAAGAAAAGAAACCGACTGGATGTGGAACCTGACCTACGGCTCAAGCTATCCGTCATCGAGCCGGATATAGATACCTTGGTGAAAGATTGCAAACAATATCATCCCTCTCACTGATATGAAACAATTATTATTATTATTATTATTTTTATAATTTANATTTTATTTTTAAAATTGTGATGAATNTTTTTAAAATTTACCTAAAATTGGTGGTCTGCGTGTGCGCCGAACGCCCATTAAGTGGTCCGCGGATGCCGAAAGTTTGAGAAACACTG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Charlie25 | AT5G61620 | 1780 | 1793 | + | 15.08 | TTGAGGATAAGATA |
| Charlie25 | DOF3.6 | 2244 | 2264 | - | 15.08 | GTTTCTTTTCTTACTCTTTAT |
| Charlie25 | SKO1 | 1903 | 1912 | + | 15.06 | ATGACGTATT |
| Charlie25 | tin | 1693 | 1701 | + | 14.91 | CTCAAGTGG |
| Charlie25 | REV | 2375 | 2388 | + | 14.90 | AATTATTATTATTA |
| Charlie25 | croc | 314 | 323 | - | 14.85 | AACGTAAACA |
| Charlie25 | POU3F3 | 328 | 340 | - | 14.81 | AAAATGCAAATTT |
| Charlie25 | ZHD6 | 145 | 156 | - | 14.73 | AATCATAATTAA |
| Charlie25 | NFIC::TLX1 | 698 | 711 | + | 14.72 | TGGCATGAAGCCAT |
| Charlie25 | CCA1 | 1086 | 1093 | - | 14.72 | AAATATCT |
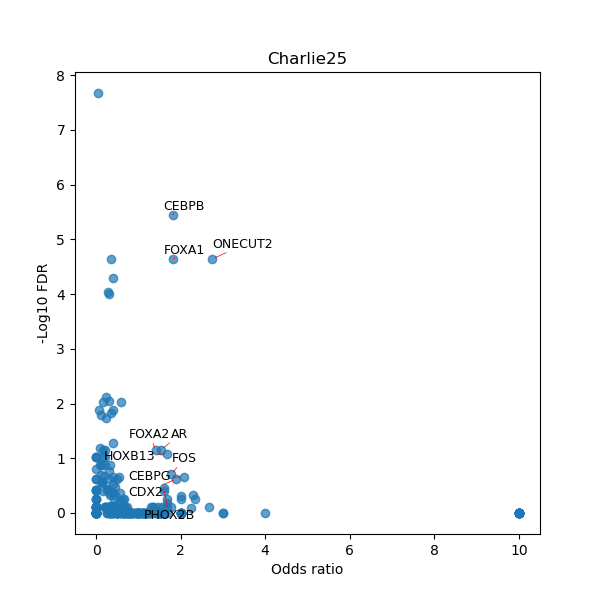
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.