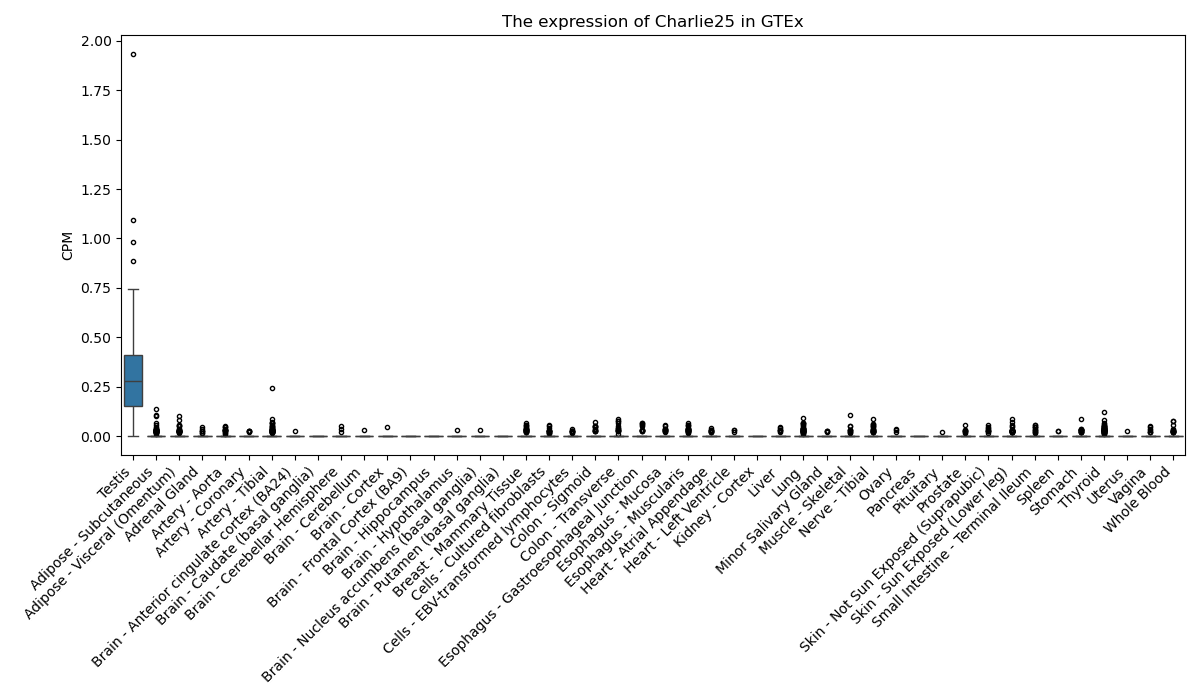
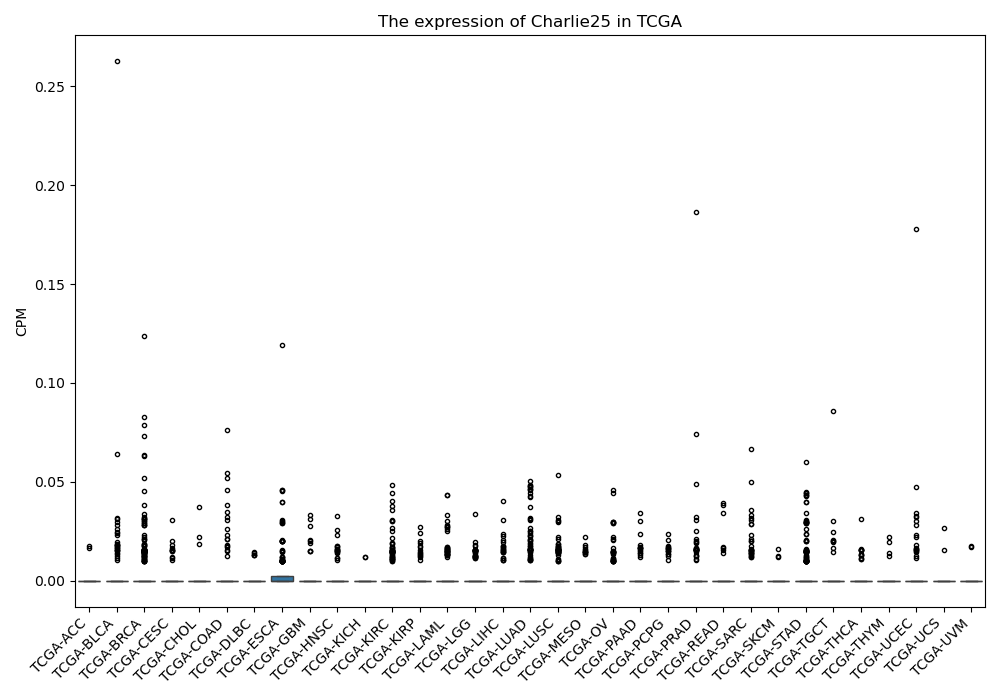
Charlie25
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000102 |
|---|---|
| TE superfamily | Charlie |
| TE class | DNA |
| Species | Eutheria |
| Length | 2524 |
| Kimura value | 30.06 |
| Tau index | 1.0000 |
| Description | hAT-Charlie DNA transposon, Charlie25 subfamily (autonomous) |
| Comment | 14 bp TIR. Complete ORF (roughly 384-2366) encodes a transposase closest to that of Charlie3 (44% identical, 62% similar). |
| Sequence |
CAGTGTTTCTCAAAGTGTGGTCCGCGGACCACTGGCGGTCCCCNGCGGTTCTATCAAGTGGTCCGCAGGCGGTTTGGCGGTTTCAGAGGAAAAAGCGATGAAACAATTTTGTTCACATACATTTCACAAATTTGAAATGTAAGATTAATTATGATTTTCACAGAAATCCCGTTACGTTCTTAATAATCGTTACGTTCTTAAAGGTTGCGCATGCGCTACAAGGACTGCGTTGGTCAGTTCGTCTCGGCTAACATTCAGTTAACAGGGTGCAGTTCGTCTCGGCTAACGTATTTTCACGTCATTTGCATGTTATTGTTTACGTTTGTTAAATTTGCATTTTTCGTTGTTACTATTGTGTTGTATTATATCCCCAATTCACAAAAATGGATCAATGGCTCAAAAGTGGTTCATTGAAGCGTAAAAGTAGTGATGAAAATAGTAACGTAAATACTACAACTCAGAATAACGTCATAAACGTAAATAGTGAACAGGACTCCAGTGCGAATATAGAATGTGAATCTGTATGTGCTGGGACAAGTGAATCTGCGAGTGTGATGATTTCGCACAAGCAGCCGAAAAAGAAAAGTGCGAATAGGAAGTACGACGATGAATATCTGAAAATTGGATTTTATTGGACCGGCGATCCATTTGCCCCTAGTCCCCAGTGCGTTGTCTGTTATGAAACTTTGTCAAATAGTGGCATGAAGCCATCGAAGCTTTCGCGTCATTTTCAAACAAAGCACAGTGACCTCTCTGGTAAACCAATCGAGTTTTTCCAGAACAAGCGCAAAATAATGCTTTCCAGTACGAAATTGATGAATTTTGTCGCTAAAGGCAGAGAAGAGACCAAAACTACAGAGGCATCATTCAAAGTTGCACTCCTTATAGCAAAAACAGGTACAAGTCACACTATTGCCGAGAAGCTTGTAAAACCAGCCGCAAAGTTAATGACAAATATTATGCTCGGAGAGAAAGCAGAACGAGCTATTGGCAAAATTCCTTTATCAAATGACACTGTTGGACGTCGCATAATATCAATGGCATACAATGTGGAAGAGCAATTACTATCACGTGTGCGTGCTAGCAGATATTTCGCTTTACAGTTGGACGAAACCACTGATGTGCAGAGTATGAATCAGCTCTTGGCATATGTGCGGTACATATATGAGGGAGAAGTGCTCGACGACTTTTTGTTCTGTTTATCACTGAAAACCCATGCTACAGGAGAAGATTTTTTTTATTTAGTTAACGATTATTTTGTGAGCCGTGATGTAGACTGGAAAAGGTGCGTTGGGATCAGTACTGACGGAGCACCAGCTATGTGTGCAGCAAGGAAGGGCGTTGCTACGCGAATAAAAGAGGTTGCACCTGAATGCCAATCCACACACTGCTTTATTCACAGAGAACAGTTGGCGGTCAACAATATGCCTCCTGATCTTGATTCAGTGTTGAAGGAAATAGTGAAAATTGTGAATACGATCAAATCGCGGCCACTGAGTGTACGTCTTTTCAGCGTGCTGTGCGAAGAAATGGGCAGCGAGTACAAGACTCTGCTTTTCCACACTGAAGTACGCTGGCTGTCGAGGGGAAAGGTGCTCACACGAGTTTTTGAAATGAGGGATGAGATAAAAACATTTCTTCATGACACTGATAATGCCAGTAAAGACCATTTCTATGATTTCAAGTGGCTTGCTCAAGTGGCGTATCTCAGCGATATATTCAGTATCTTGAATAGCCTGAACCTATCACTTCAGGGCCGAAATATCACGATTTTTAATGTTGAGGATAAGATATCAGGATTTCTTAAGAAGACCGAACTGTGGTGCAAACGGCTCGATCGCCGAGAGTTTGACTCTTTCCCAACACTTGATGATTTTCTTCACTCGTCGGAGAAAGAAATCGATGACGTATTATTGGGCATATTTAAAAACCACATCCAAATGCTGCAACAGAACATGAAGAAATACTTTCCAGAGCCGAATGCAACCAAAGAGTGGATTAGGAATCCATTCGCCGCTATCTCCCAAGTTGAAACATTCAACCTTCCAGCTTTTGAGTGTGATATGCTCGTCGACTTAGCGTCTGACGGAGCGCTGAAAGTAGTCTTCAGTGAGAAATCTCTCCATAATTTCTGGGTCCATGTTCGATCCGAATATCCAGAACTATCTGACAGAGCCACCAAACACTTGCTGCCATTCCCGACGACCTATAATTGTGAGTTAGGATTTTCTAATTTAGTAGAAATAAAGAGTAAGAAAAGAAACCGACTGGATGTGGAACCTGACCTACGGCTCAAGCTATCCGTCATCGAGCCGGATATAGATACCTTGGTGAAAGATTGCAAACAATATCATCCCTCTCACTGATATGAAACAATTATTATTATTATTATTATTTTTATAATTTANATTTTATTTTTAAAATTGTGATGAATNTTTTTAAAATTTACCTAAAATTGGTGGTCTGCGTGTGCGCCGAACGCCCATTAAGTGGTCCGCGGATGCCGAAAGTTTGAGAAACACTG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Charlie25 | FKH1 | 312 | 324 | - | 16.08 | AAACGTAAACAAT |
| Charlie25 | DOF5.1 | 576 | 594 | + | 15.97 | AAAAAGAAAAGTGCGAATA |
| Charlie25 | F10B5.3 | 772 | 782 | + | 15.84 | TTTTCCAGAAC |
| Charlie25 | NFIX | 698 | 711 | - | 15.83 | ATGGCTTCATGCCA |
| Charlie25 | TCX6 | 123 | 137 | - | 15.73 | TTTCAAATTTGTGAA |
| Charlie25 | HOXB13 | 627 | 635 | - | 15.62 | CCAATAAAA |
| Charlie25 | ZBTB6 | 2290 | 2298 | - | 15.31 | GCTTGAGCC |
| Charlie25 | FOXF2 | 313 | 321 | - | 15.28 | CGTAAACAA |
| Charlie25 | FOXN3 | 313 | 320 | - | 15.19 | GTAAACAA |
| Charlie25 | ceh-22 | 1680 | 1688 | - | 15.16 | CCACTTGAA |
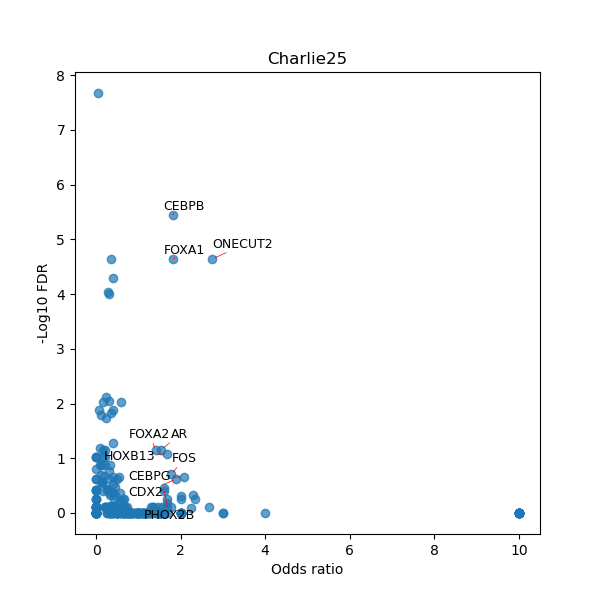
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.