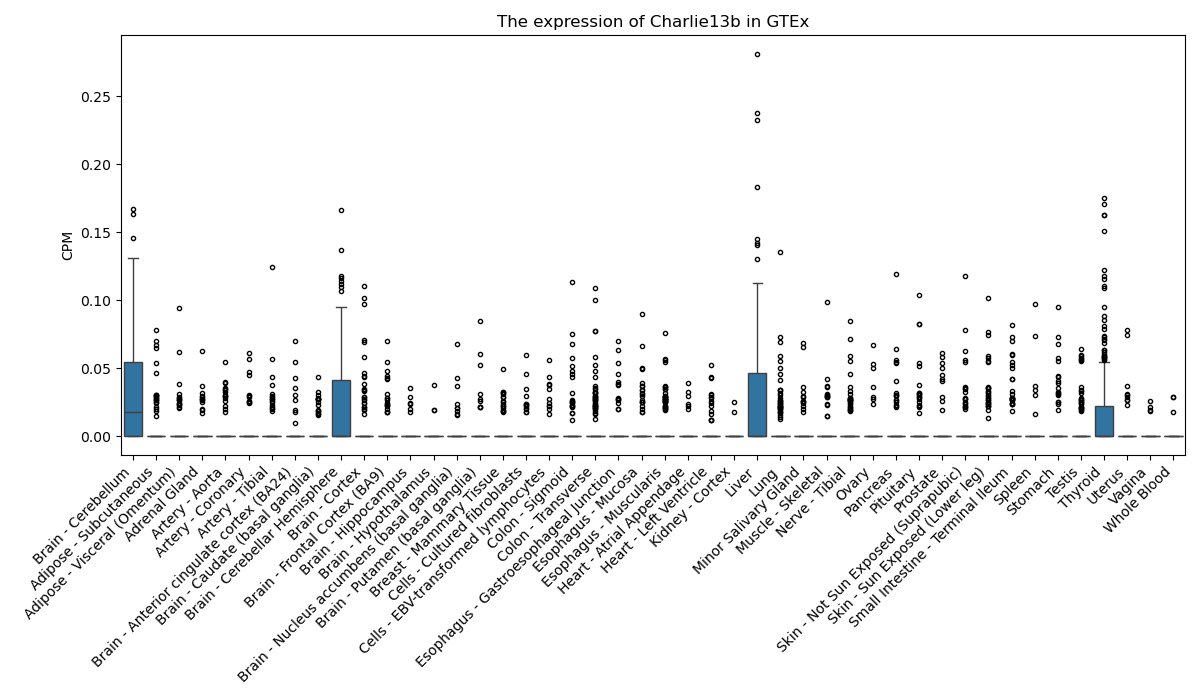
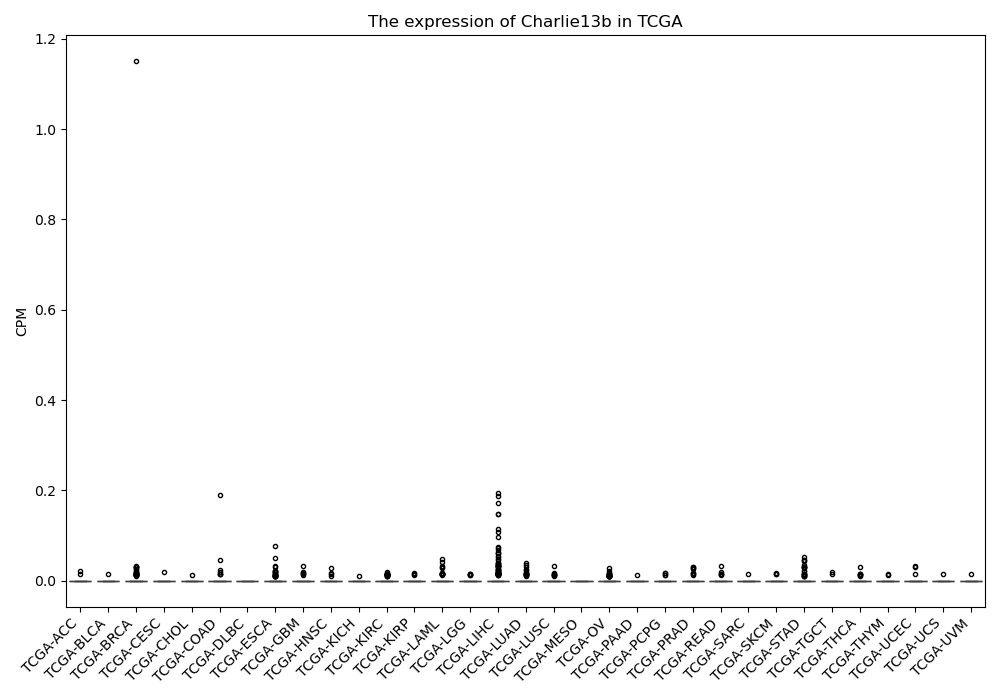
Charlie13b
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000087 |
|---|---|
| TE superfamily | Charlie |
| TE class | DNA |
| Species | Mammalia |
| Length | 512 |
| Kimura value | 31.78 |
| Tau index | 1.0000 |
| Description | hAT-Charlie DNA transposon, Charlie13b subfamily (non-autonomous) |
| Comment | Vague 8bp TSDs flank the current ends. |
| Sequence |
CANGGGCATCCAACATGCGGCCCGCGGGCCAAATGCGGCCCGCCTGACCCCAGGGTGCGGCCCGCCTGAGATTTTTAGCAAAAATGTTTTTACCTAATTAGCTAGCACACATGAACTAGTAGGGCCAGNAGGGCCTAACCACCTGCCCGCCGCACCACTGTGTAACTGACTGCCGCCAGGTGTCATGCTAGCAGGCTTGGATCAATTAGTAATTGTAATTGAAGCTGAATGTAATTAATTATATAATTTTGAGCTGTTCAAGTTTAATTCCACTAAAATTACATCATGAAACTGGGGCATTATGAAGGATTACTGACAAAAGGAAAATAGGTATGAATAAAGCCTCCATATGCACTTGACTCACGTTTGAGTCACGTCCAATACTCGTACGCCAAGCGTGGACCCTCTCGACTCACCATTGTTATCAATAAAAAAATTCATGCGGCCCGCACACATATGGATTTCTGATCATGTGGCCCACTATTGAAAAAACNTGGACGCCCCTGCCCTAG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Charlie13b | MYB39 | 89 | 100 | + | 14.10 | TTTACCTAATTA |
| Charlie13b | CEBPG | 278 | 287 | - | 14.07 | ATGATGTAAT |
| Charlie13b | ato | 139 | 147 | + | 14.07 | CCACCTGCC |
| Charlie13b | CIN5 | 238 | 247 | + | 14.05 | ATTATATAAT |
| Charlie13b | CIN5 | 238 | 247 | - | 14.05 | ATTATATAAT |
| Charlie13b | usp | 43 | 51 | - | 14.00 | GGGGTCAGG |
| Charlie13b | KLF3 | 53 | 62 | - | 13.98 | GGCCGCACCC |
| Charlie13b | MYB41 | 90 | 99 | + | 13.93 | TTACCTAATT |
| Charlie13b | ZNF454 | 14 | 30 | - | 13.87 | GGCCCGCGGGCCGCATG |
| Charlie13b | Lhx1 | 93 | 102 | - | 13.80 | GCTAATTAGG |
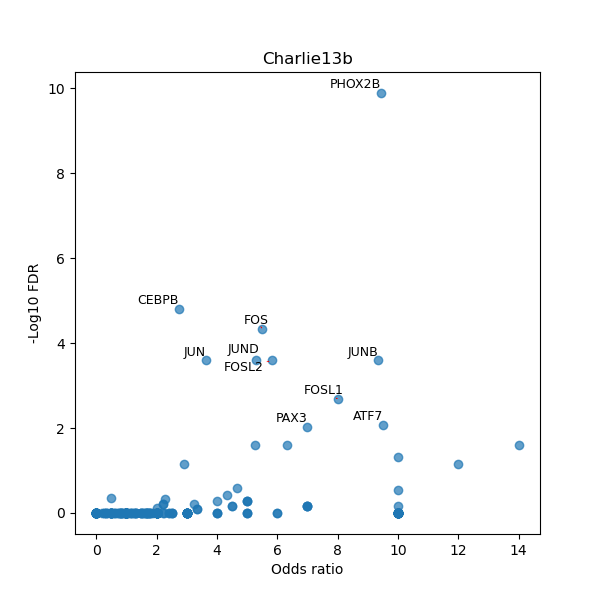
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.