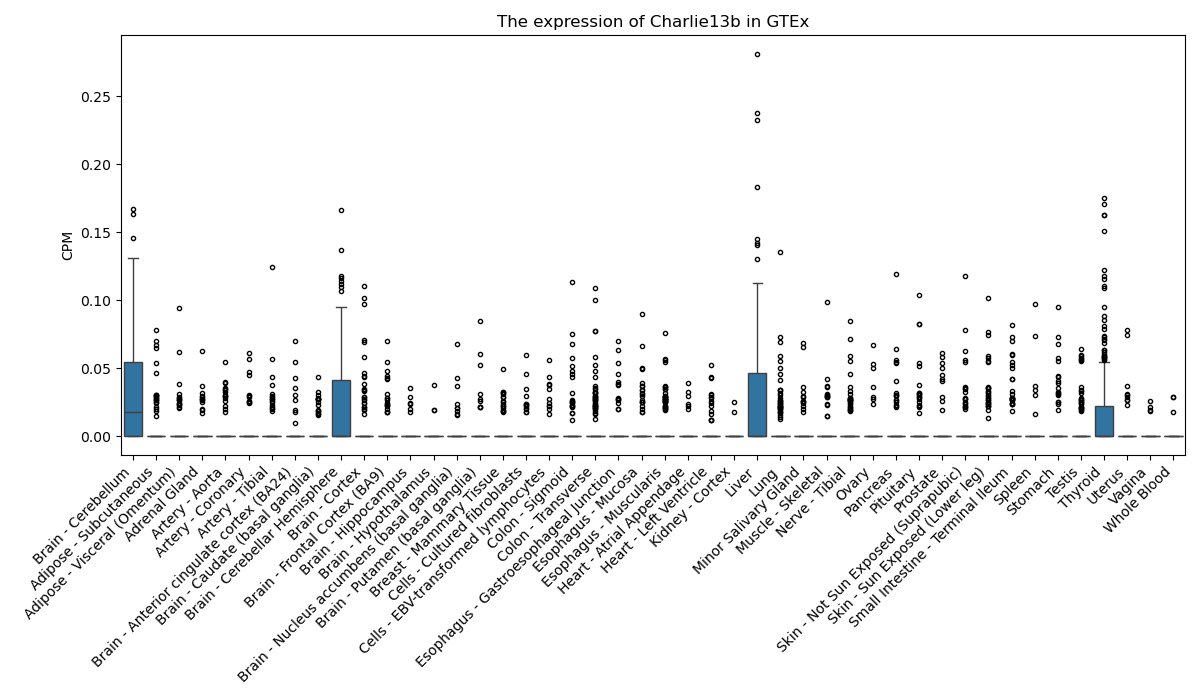
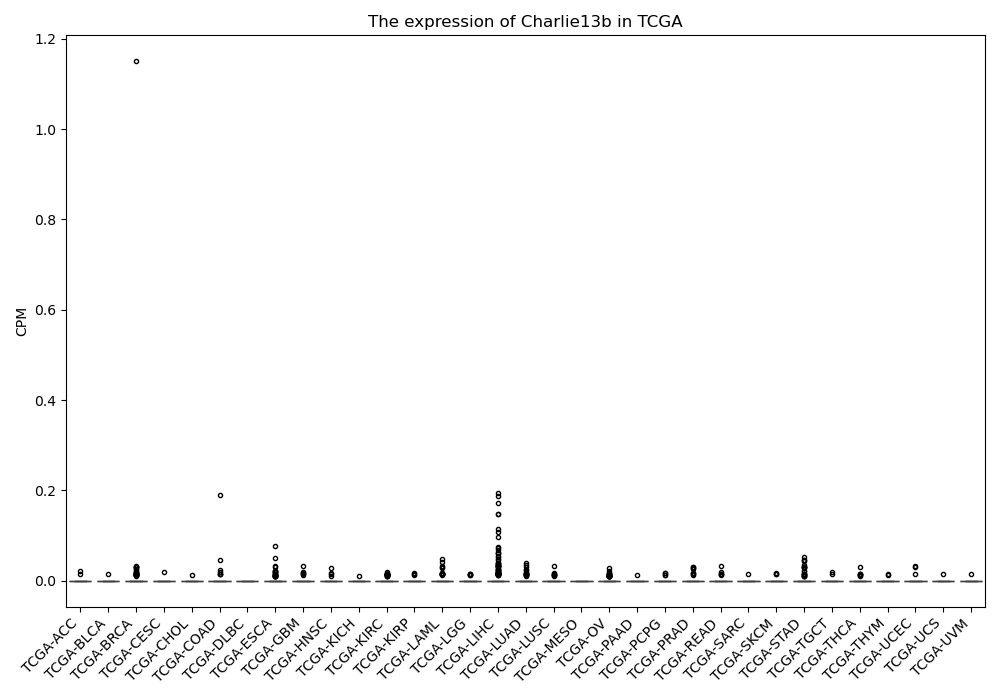
Charlie13b
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000087 |
|---|---|
| TE superfamily | Charlie |
| TE class | DNA |
| Species | Mammalia |
| Length | 512 |
| Kimura value | 31.78 |
| Tau index | 1.0000 |
| Description | hAT-Charlie DNA transposon, Charlie13b subfamily (non-autonomous) |
| Comment | Vague 8bp TSDs flank the current ends. |
| Sequence |
CANGGGCATCCAACATGCGGCCCGCGGGCCAAATGCGGCCCGCCTGACCCCAGGGTGCGGCCCGCCTGAGATTTTTAGCAAAAATGTTTTTACCTAATTAGCTAGCACACATGAACTAGTAGGGCCAGNAGGGCCTAACCACCTGCCCGCCGCACCACTGTGTAACTGACTGCCGCCAGGTGTCATGCTAGCAGGCTTGGATCAATTAGTAATTGTAATTGAAGCTGAATGTAATTAATTATATAATTTTGAGCTGTTCAAGTTTAATTCCACTAAAATTACATCATGAAACTGGGGCATTATGAAGGATTACTGACAAAAGGAAAATAGGTATGAATAAAGCCTCCATATGCACTTGACTCACGTTTGAGTCACGTCCAATACTCGTACGCCAAGCGTGGACCCTCTCGACTCACCATTGTTATCAATAAAAAAATTCATGCGGCCCGCACACATATGGATTTCTGATCATGTGGCCCACTATTGAAAAAACNTGGACGCCCCTGCCCTAG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Charlie13b | ATF4 | 278 | 287 | - | 15.09 | ATGATGTAAT |
| Charlie13b | ASCL1 | 139 | 147 | + | 14.74 | CCACCTGCC |
| Charlie13b | PAX3 | 232 | 241 | + | 14.72 | TAATTAATTA |
| Charlie13b | PAX3 | 232 | 241 | - | 14.72 | TAATTAATTA |
| Charlie13b | Sox17 | 416 | 425 | - | 14.61 | ATAACAATGG |
| Charlie13b | kni | 289 | 299 | + | 14.48 | AAACTGGGGCA |
| Charlie13b | ZmbZIP57 | 278 | 287 | - | 14.40 | ATGATGTAAT |
| Charlie13b | Sox5 | 416 | 423 | - | 14.38 | AACAATGG |
| Charlie13b | Pax7 | 232 | 241 | + | 14.36 | TAATTAATTA |
| Charlie13b | Pax7 | 232 | 241 | - | 14.36 | TAATTAATTA |
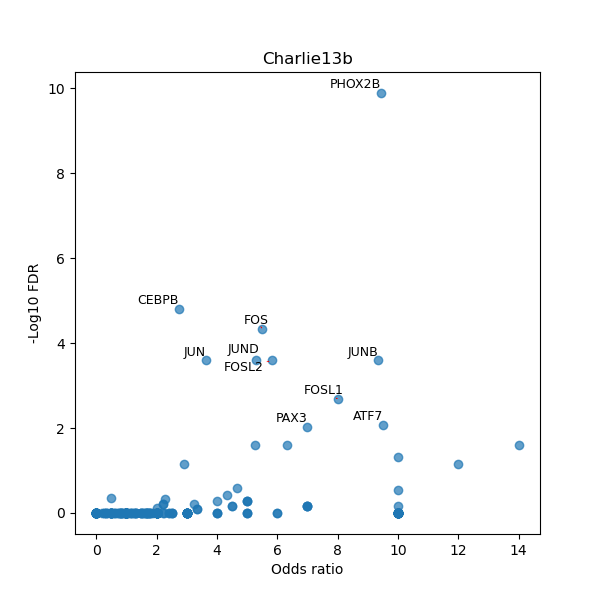
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.