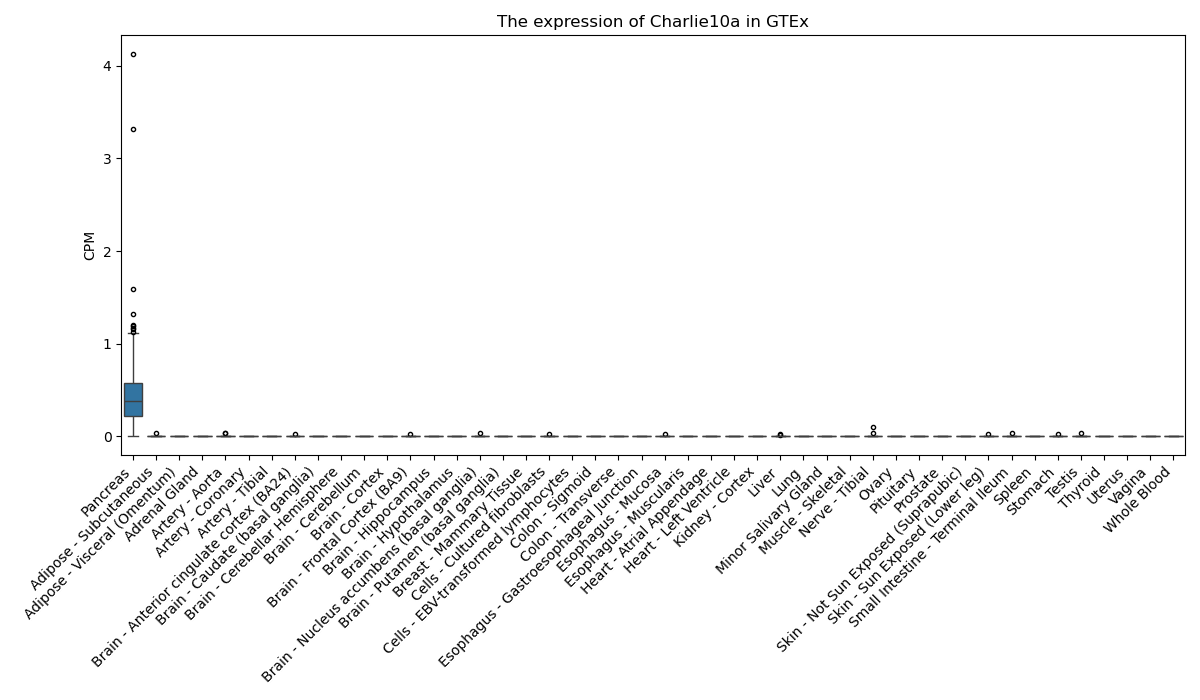
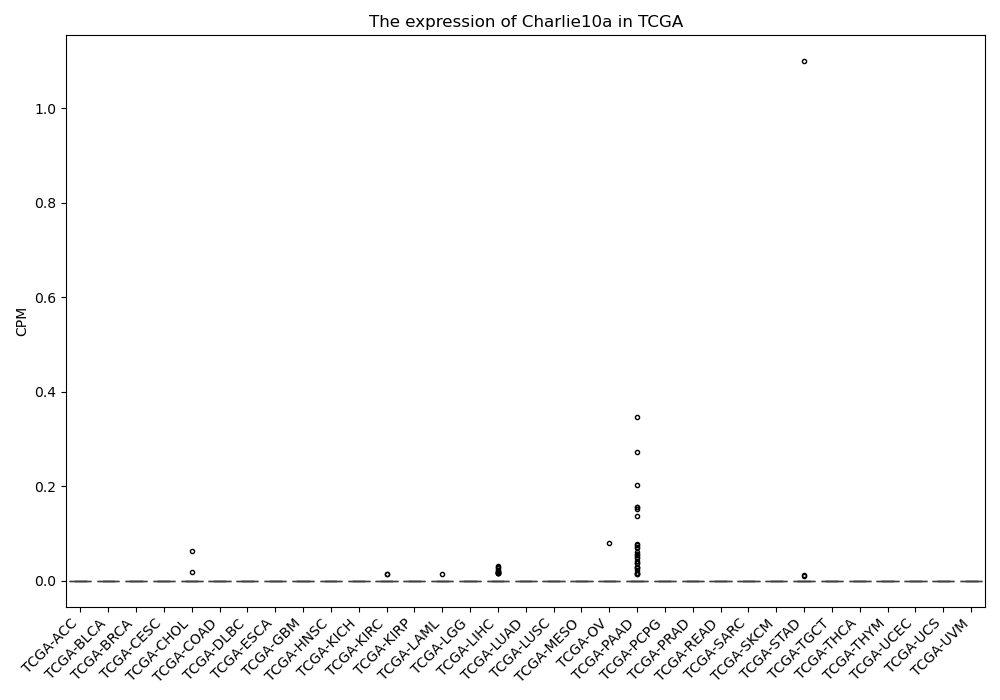
Charlie10a
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000082 |
|---|---|
| TE superfamily | Charlie |
| TE class | DNA |
| Species | Eutheria |
| Length | 280 |
| Kimura value | 23.06 |
| Tau index | 1.0000 |
| Description | hAT-Charlie DNA transposon, Charlie10a subfamily (non-autonomous) |
| Comment | - |
| Sequence |
CAGTGCTACTCAAAGTGTGGCCCATGGACCGGTGCCAGTCCGCGAACTGTTTGTTACCGGTCCGCGACGAGATAAGGAGCTTGCGCCAGAATGTAAATCAACGCACTGCTTCCTTCATCGAGAAAGTCTTGCTACGAAAAAAATGTCAGCTGAACTAAACAGTGTGCTTAGTGACGTAGTTGATTTACATTCTGGCGCAAGCTCCTTATCTCGTCGCGGACCGGTAACAAACAGTTCGCGGACCGGCACTGGTCCGCGGACCACACTTTGAGTAGCACTG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Charlie10a | SRM1 | 70 | 78 | + | 13.66 | AGATAAGGA |
| Charlie10a | PRDM1 | 120 | 126 | - | 13.28 | CTTTCTC |
| Charlie10a | NFIC | 24 | 38 | + | 13.19 | ATGGACCGGTGCCAG |
| Charlie10a | srp | 204 | 211 | - | 13.11 | AGATAAGG |
| Charlie10a | srp | 70 | 77 | + | 13.11 | AGATAAGG |
| Charlie10a | bZIP14 | 171 | 178 | + | 13.09 | GTGACGTA |
| Charlie10a | Atf1 | 171 | 178 | + | 13.08 | GTGACGTA |
| Charlie10a | ZNF320 | 15 | 34 | + | 12.93 | GTGTGGCCCATGGACCGGTG |
| Charlie10a | GATA2 | 205 | 211 | + | 12.60 | CTTATCT |
| Charlie10a | GATA2 | 70 | 76 | - | 12.60 | CTTATCT |
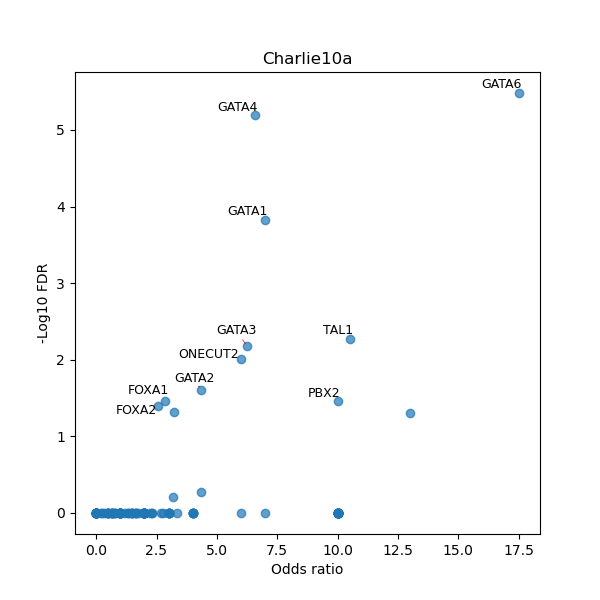
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.