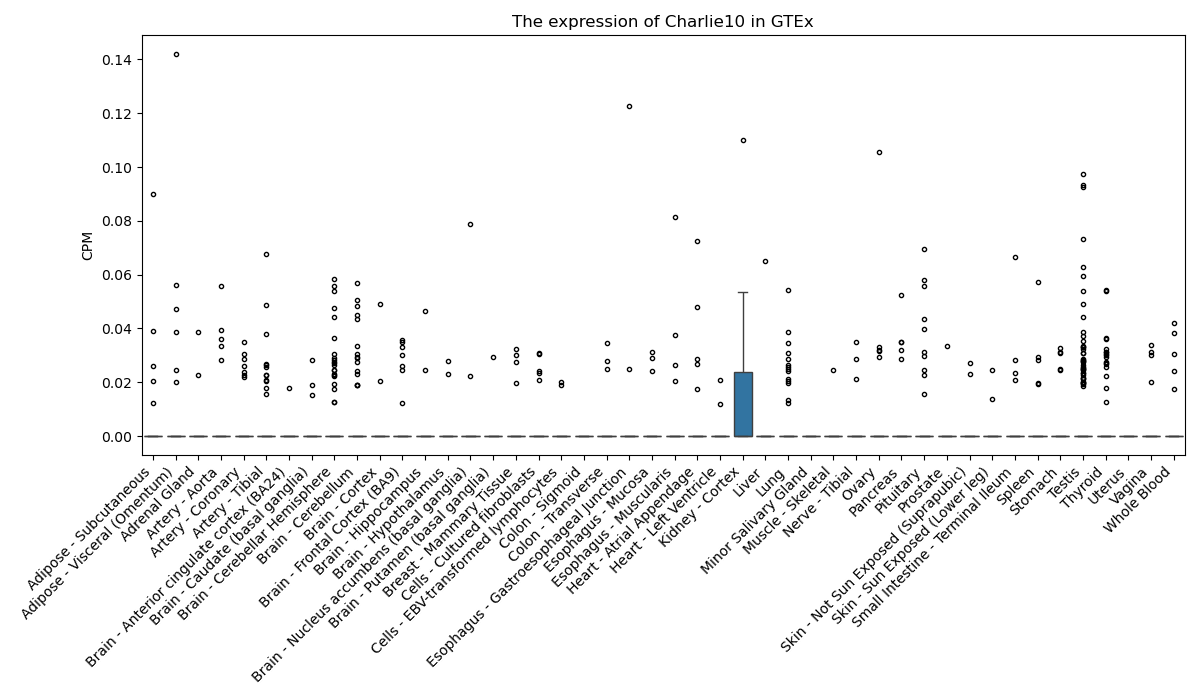
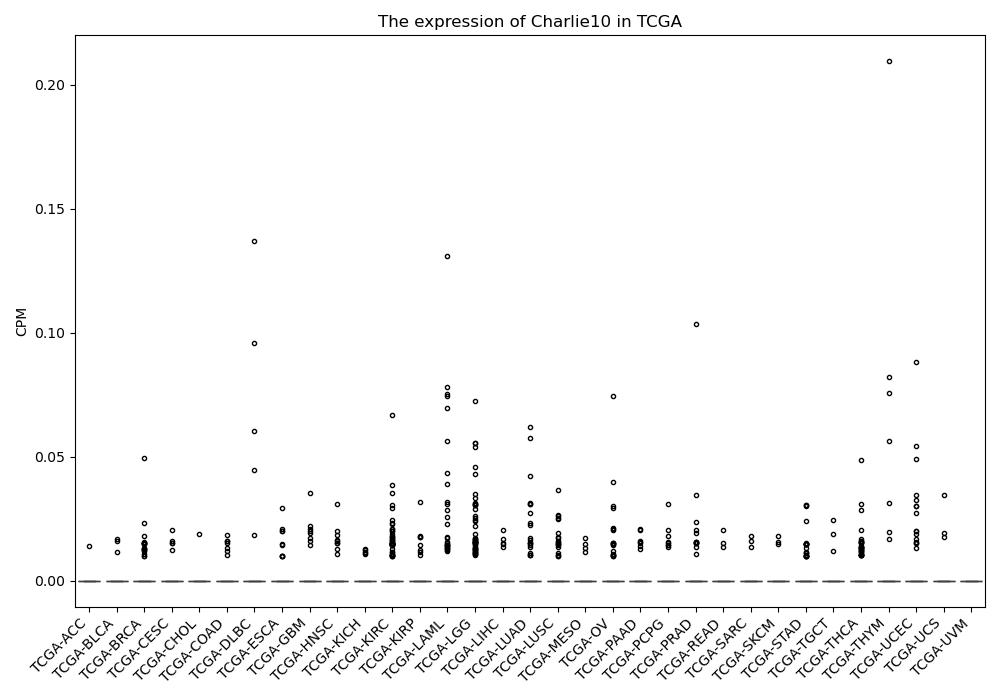
Charlie10
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000081 |
|---|---|
| TE superfamily | Charlie |
| TE class | DNA |
| Species | Eutheria |
| Length | 2822 |
| Kimura value | 24.12 |
| Tau index | 0.0000 |
| Description | hAT-Charlie DNA transposon, Charlie10 subfamily (autonomous) |
| Comment | Charlie10 is a full-length hAT-Charlie DNA transposon. MER5C is an internal deletion product of Charlie10. MER5A and B, with 65000 copies by far the most numerous DNA transposon remnants in the human genome, have closely matching termini, but a dissimilar (short) internal region. Charlie10 may have caused their distribution, in analogy to the transposition of Ds1 by Activator. The human gene KIAA1923 (WDR59) appears to be an ancient, domesticated Charlie10 transposase. |
| Sequence |
CAGTGCTACTCAAAGTGTGGTCCGTGGACCGGTGCCGGTCTGCGAACTGTTTGTTACCGGTCCATGATGAGATAAGTACAGAAATTGAGAGTAAGCGTTTAGAAACTTTTATAGCAATTTGACATTGCCGCGACATCCAAGCGCGTGATCAGTGGACTCGTCTCGTTGAACAGGGTATAGACCAGTTCGGGTGTTGTCGAACTCGCGTGGTGAGTCGCATGTGGCGCGAGCTGCGTACTAGTCATGCGCAGTAGGACCACGTTACAACGTGTGATATTTTAAGGTTAGTTTGTCAACTGTAACCCAAAAGTATATAAAAATCTGAGAAAATGTAAGCATTTATTTATTTTTATAACTTATTTATTTTTATAATCANTTTAANTTTTAATCAAGCCTAGTTTTTAATTTATAGTTTAACATTATTAGCTAAATTAGGGAAGTAAAATTTGTATTATTTATTTTTAACCCTAATTTACNGATGGCGAAACAAGCTTCACTGGACTTTTTTGTCAAGAAAAGACACGCCTTTTCTGAATGCGGCAGTAGTAGTAAACCAAATGATGACGATGGAAGTGATTCCGAAGAAATGAAGACCAAAAGAGTTCGTACTAGTTTTACTCGGAAGTATGATCCCTCATATATTGAGTTCGGTTTTGTAGCCATAATTGATGGTGAAGTGCTAAAACCACAGTGTGTTATTTGCGGAGATGTACTGGCTAATGAAGCAATGAAACCATCAAAACTTAAGCGGCATTTACATACAAAACATAAAGAAATAAGTTCAAAACCAAAAGAATTNTTTGAAAGAAAGAGTATTGAATTAAAAAGCCGACAGAAGCAGATGTTCAATATTTCACATATAAACATTAGTGCTTTGCGGGCTTCTTATAAAGTAGCACTTCGGGTTGCTAAGACTAAAANACCATATACAATTGCCGAGACATTAGTGAAAGACTGCATCAAAGATGTTTGCTTGGAAATGTTGGGTGAATCTGCGGCAAAGAAGGTAGCTCAAGTACCACTTTCCAATGACACCATAGCTCGACGTATTCAGGAACTGGCTAATGATATGGAAGACCAACTCACAGAACAAATAAAGCTAGCAAAGTATTTTTCATTGCAACTTGACGAATGCACAGATATTGCTAACATGGCAATTCTTTTAGTATATGTGCAATTTGAACATGATGGTGATATGAAGGAAGAATTCTTTTTTTCAGCTTCATTGCCGACAAACACAACTAGCTCTGAACTGTATAAAACTATGAAGGATTACATTGTCAACAAATGTGGTTTGGAGTTTAAGTTTTGTGTAGGAGTATGTTCTGATGGCGCAGCTGCAATGACAGGAAAACATTCTGGAGTAGTTACCCAGATTAAGGAGCTTGCGCCAGAATGTAAATCAACGCACTGCTTCCTTCATCGAGAAAGTCTTGCTACGAAAAAAATGTCAGCTGAACTAAACAGTGTGCTTAGTGACGTAGTAAAAATTGTGAATTACGTAAAGGCTAATGCGTTAAATTCGAGATTATTCTCTTTATTATGTGATAATATGGAAGCTGATCATAAACAACTGTTATTGCATGCTGAGGTACGATGGTTATCGAGGGGAAAAGTTCTGTCGAGAATGTTTGAACTACGGAACGAACTCTTAGTGTTTCTGCAAGATAAGAAACCAGTTTGGTCCCAACTTTTTAAAGATGTGAATTGGACAGCCAGACTTGCTTATTTGTCTGATATCTTCGGTATTTTTAATGATCTTAATACTTCCATGCAAGGAAGGAATGCAACATGTTTTTCAATGGCAGATAAGATCGAAGGGCAAAAACGAAAGTTAGAAGCTTGGAAGAACAGAGTTTCTACAGATTGTTATGACATGTTCCATAATTTAACAACAATTATCAATGAAGTAGGTGATGATCTTGATATTGCACATCTGCGAAAAGTTATCACCGAACACCTTACGAATTTGATAGAACGTTTTGAATTTTATTTTCCATCAAAAGAAGATCCACGCATAGGAAATTCATGGATCCGGAATCCATTTCTTTCATCGAAAGATAATTTAAATTTAACTATAACTTTACAGGATAAATTGTTGGAACTGGCTACTGATGAAGGATTGAAGATGAATTTTGAAAATACAGCATCACTTGCTTCATTTTGGATAAAAGTTAAAAATGAATATCCTGAGCTTGCTGAAATTGCTTTAAAATCTCTTCTTCCATTCCCGTCAACATACCTCTGTGAGACTGGTTTCTCTACTATGAGTGTTATTAAAACAAAACATAGAAACAGTTTAGATATACATTATCCCCTGCGAGTAGCGTTGTCATCAATCCAACCTAGATTAGATAAGTTAACAAGCAAGAAGCAAGCTCATTTGTCACATTAAAAACTTTAAATATTGATATACACGGTGTTCGTTCAAAGTGTGCATATAGGCGTTTAATATGGGGAATCGCTATTTCACTTCATAACTTTTTGTTTAGTTATAATTGCGGAACGACAAAAAGTTACGAGTGTAATAGCGATTCTCTATATTAATCGCCTATATGCACACTTTCAATGAACAATGTGTACAGTTTTTGTAATTCTTTTTTTCCTTATATTTGTTATATTTATTGAAATGTAATTTTATGTCTGTTGAATCTAATAATAAAAAATTTTGGGCTTGTATTTTGTATGTCTTTTTTTTTTAAATTTCATTTTTCTAGTAATTCATTTTTATTGTATTTTACAAAAGTATCGGTCTGCGATAGATTGGAAATTAAAAAAAAAAAAAAACTGGTCCTTCACCACAGATAGTTTGAGAAGCACTG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Charlie10 | ceh-38 | 338 | 351 | - | 14.66 | AAAAATAAATAAAT |
| Charlie10 | MYF6 | 1569 | 1578 | - | 14.64 | AACAGTTGTT |
| Charlie10 | ARF18 | 1229 | 1236 | + | 14.63 | CCGACAAA |
| Charlie10 | AT5G61620 | 2162 | 2175 | + | 14.59 | TTTTGGATAAAAGT |
| Charlie10 | GATA1::TAL1 | 1655 | 1671 | - | 14.55 | TTATCTTGCAGAAACAC |
| Charlie10 | ac | 1334 | 1341 | + | 14.47 | GCAGCTGC |
| Charlie10 | ac | 1334 | 1341 | - | 14.47 | GCAGCTGC |
| Charlie10 | TCX2 | 2697 | 2711 | - | 14.46 | AAATGAAATTTAAAA |
| Charlie10 | GRHL1 | 1673 | 1682 | + | 14.44 | AAACCAGTTT |
| Charlie10 | TBF1 | 464 | 471 | + | 14.43 | AACCCTAA |
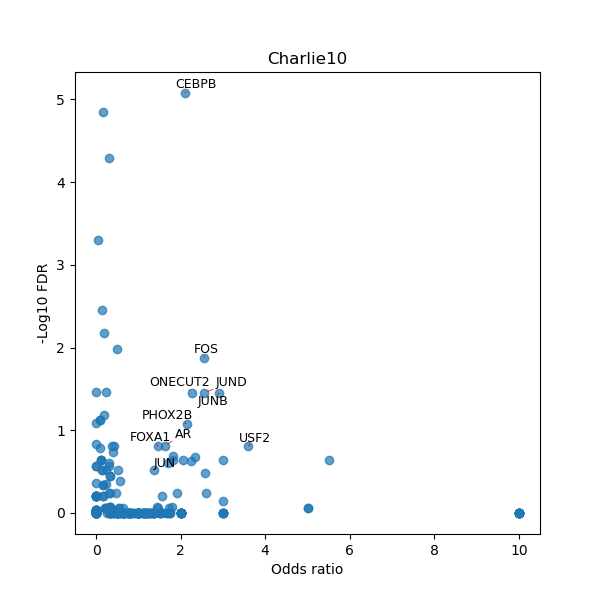
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.