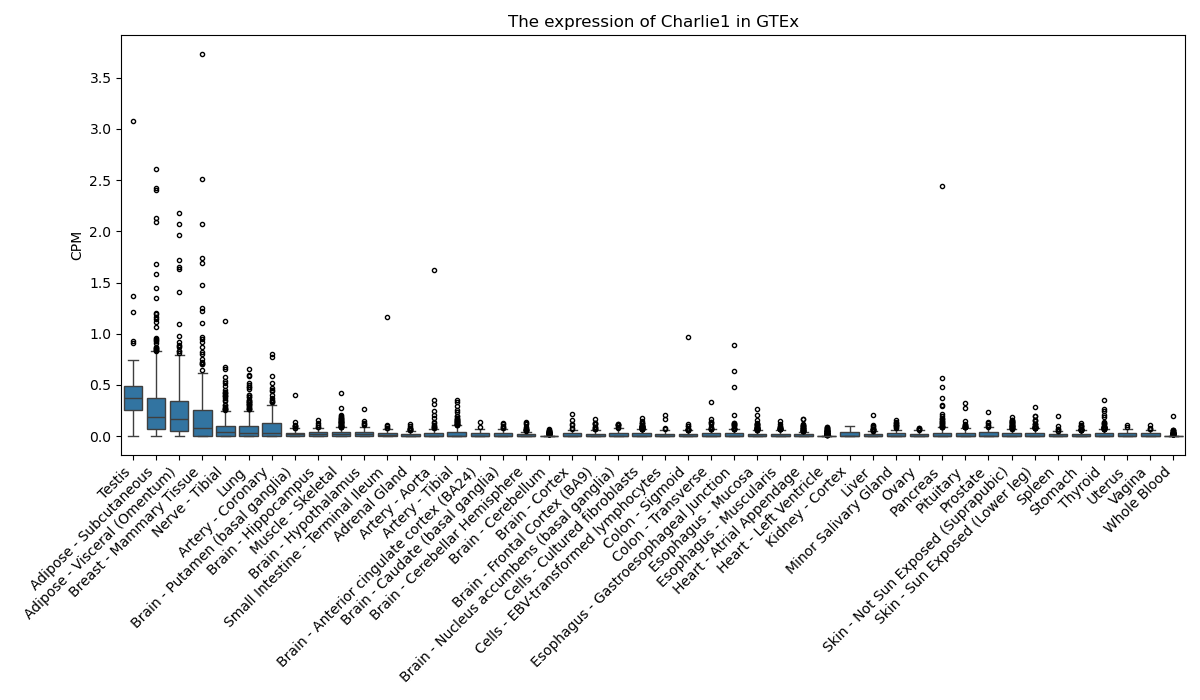
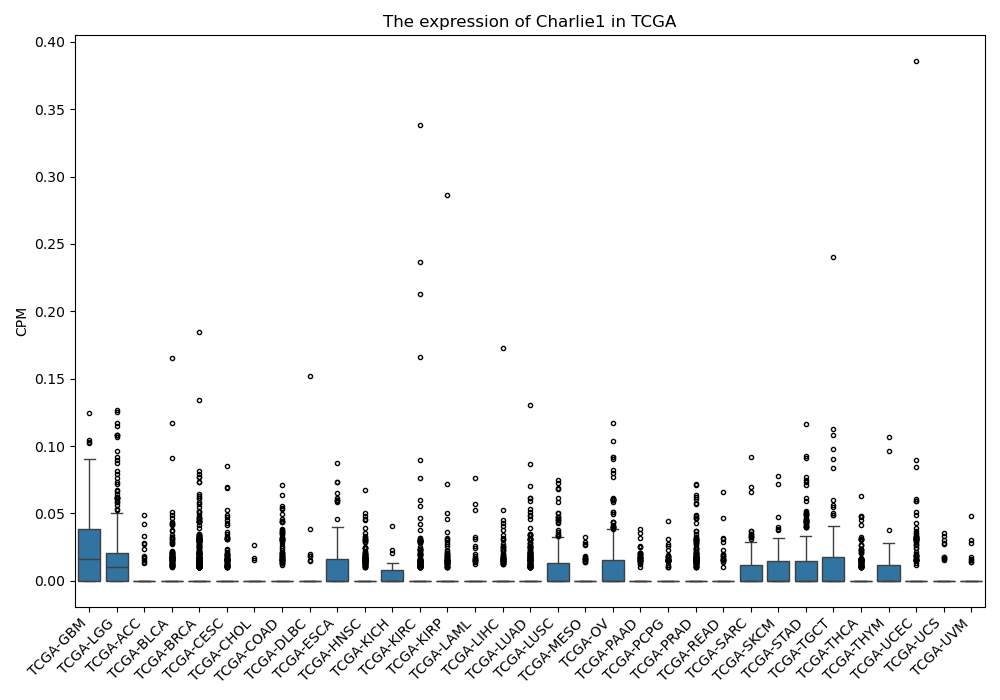
Charlie1
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000018 |
|---|---|
| TE superfamily | Charlie |
| TE class | DNA |
| Species | Metatheria,Eutheria |
| Length | 2781 |
| Kimura value | 20.20 |
| Tau index | 0.9634 |
| Description | hAT-Charlie DNA transposon, Charlie1 subfamily (autonomous) |
| Comment | 15-16 bp terminal inverted repeats. 8 bp TSD. |
| Sequence |
CAGCGGTTCTCAAAGTGTGGTCCGGGGACCCCTGGGGGTCCCCGAGACCCTTTCAGGGGGTCCGCGAGGTCAAAACTATTTTCATAATAATACTAAGACGTTATTTGCCTTTTTCACTCTCATTCTCTCACGAGTGTACAGTGGAGTTTTCCAGAGGCTACATGACGTGTGATGACGTCATCGCTCTGACGGCTAATGGAATGTGTGCTTGTGTATTCTTGTGTTTTCTAGAATTTTCTAAGGTAGTAGGTTTAGGGTATAAATATGTGCGTTTTCAGAGATTAACTCAGTTTGTTCTCAGTACTTCTACTGTGCTCTTACTAGCTATCTTCGGTTATACCTGCTATAATCTCTGTAACCTCATTATCGTCCAATAAATCGTTATTTTGAAATCCCGAAGTTTTCCTTGTGCCTACGCGGAAACACAACAAGAAGTAAGTACACTTTGTTGTCTTGTTTTGCAATAATATTTTNAAATTTTTTGAAAACTTTTCTAAATTTTAAAATTTTATCTAAAACTATTATTTTAGAATTTAATAATATTTATTCTAAAATAAAAGAAAATTTATTTATAATATATTTTTCTTATATTTAAGGATGGATCGTTGGCTTNAAAAAGGGAGATTAGAAGACTTGCTTTCTCAACCCACAGCTGCACCGTCTACGTCTAAAGATGCTGAAAAAGATGAAACTGACATATCAGAGAGTTCTCTGACTCATGGAAGGGAGGAATCTACTCCAAAGAAACTGGGCGAAACTGTAAATAAGAAACGAAAATATGATGAAAGCTATCTTTCCCTCGGCTTTATAGATGTTAATAATTTACCTTATTGTGTCTTATGCAACAGAACATTTTCGAATAGTATTATGGTGCCAGTTAAGTTGCGGCATCATTTTGAGACCAATCATTCAGAGTTTAAAGAAAAAGGAATTGAATATTTTAAACGTAGACGTGATGAGCTCTTTAAAAGCCAAAAATTGTTTGTTACAGCTTTTCAAACTAGAAATGAAAAAGCCACTGAAGCATCTTACAGGGTAAGTTATCGTATTGCATTGGCTGGAGAAGCGCACACAATAGCTGAGAGACTAATAAAGCCTTGTACAGTTGACATTGCTGAATGCCTGCTGGATGAAAAGTCAGTAAAAGAAATCACGGCACTGCCACTTTCCAATGATACAGTAACTCGTCGAATTAAAGATTTAGCTGCAAACATGAAGACTGAGTTAATATCTCGTCTGCAGAATTGTACTTTTGCCTTACAAATGGACGAATCTACAGACGTGGCTGGACTTGCTGTTTTGCTTGTATTCGTCCGGTATCAGCACCAACTAATCATCGAAGAAGATCTTTTATGTGAATGCTTGGCAACAAACACAAGTGGTGCTGAAATATTCAAAGTGTTGAATAACTTTTTTGAATCTCATGGTTTATCCTGGAACAACTGTGTTGACATTTGCACTGATGGTGCAAAAGCAATGGTGGGTAAAACTGCTGGCGCCTTAGCACGAATCAAGGCAGTGGCACCAAACTGTACTAGTAGTCATTGTATTCTTCACCGCCACGCACTCGCAGTNAAAAAAAAGCCAGTTTCACTTAAGAATGTCCTTGATGAAGCAGTAAAAATTATTAATTTTATTAAATCTCGACCCTTGAGTACACGTCTTTTTAATATTCTGTGTGACGAAATGGGAAGTACGCATAAAGCACTTCTGCTGCATACCGAAGTACGATGGTTGTCTCGAGGAAAAGCACTTGTGCGATTGTTTGAGTTGCGAGCTGAACTAGCCGCTTTTTTCATGGAACACCATTTTTACTTGAAAGAACGACTGACAGACAAACTATGGTTATTCAGACTTGGGTATTTGGCAGACATTTTCTCGAAAATGAACGAAGTGAGCCTGTCACTTCAAGGAAAACAACTGACAGTATTTGTTGCCAATGATAAAATTCGAGCTTTCAAGCGAAAATTAGAATTTTGGAAAACTTGTATCCGCCACCGTGAGCTTGACAGCTTCCCAATACTTAAAGACTTTTCTGATGAGATCGGTGGTGATATTAACGAATGTGATTTTTTGATATTGTATAATGAAATGTGTCAACATTTGGAAGATCTGCATAACTCAGTGAACCAATATTTTCCAAATGACCAATGCATGATGTTACAAAATCATGCATGGGTAAAAGATCCATTCAAAGTGCAAGATAGACCAATGGATTTTAATGTAACAGAGTACGAAAAGTTCATTGATATGGTTTCAGATTCCACATTGCAACTAACCTTTAAGAAACTACCACTTGTCGAGTTTTGGTGTAGTATCAAAGAAGAATATCCACAATTATCTGAAAAGGCTATTAAAATACTCCTCCCTTTTCCAACTACATATCTGTGTGAGGCCGGATTTTCTTCATATACTTCAACCAAAACAACATATCGCAACAGATTGAATGCAGAAGCAGATATGAGAATCCAGCTGTCTTCTATTAAGCCAGACATTAAAGAGATTTGCAAAAATGTAAAACAATGCCACTCTTCTCACTAAATTTTTTTGTTTTGGAAAATATAGTTATTTTTCATAAAAATATGTTATTTATGTTAACATGTAATGGGTTTATTATTATTTTTAAATGAATTAATAAATATTTTAAAAATTTCTCAGTTTTAATTTCTAATACGGTAAATATCGATAGATATAACCCACATAAACAAAAGCTCTTTGGGGTCCTCAATAATTTTTAAGAGTGTAAAGGGGTCCTGAGACCAAAAAGTTTGAGAACCGCTG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Charlie1 | TGA9 | 171 | 181 | + | 19.08 | GATGACGTCAT |
| Charlie1 | TGA9 | 172 | 182 | - | 19.08 | GATGACGTCAT |
| Charlie1 | HSFA6A | 226 | 238 | - | 18.85 | GAAAATTCTAGAA |
| Charlie1 | Atf-2 | 171 | 181 | + | 18.80 | GATGACGTCAT |
| Charlie1 | Atf-2 | 172 | 182 | - | 18.80 | GATGACGTCAT |
| Charlie1 | Fos-b | 172 | 181 | + | 18.67 | ATGACGTCAT |
| Charlie1 | Fos-b | 172 | 181 | - | 18.67 | ATGACGTCAT |
| Charlie1 | ATF7 | 172 | 181 | + | 18.64 | ATGACGTCAT |
| Charlie1 | ATF7 | 172 | 181 | - | 18.64 | ATGACGTCAT |
| Charlie1 | TGA4 | 170 | 180 | - | 18.45 | TGACGTCATCA |
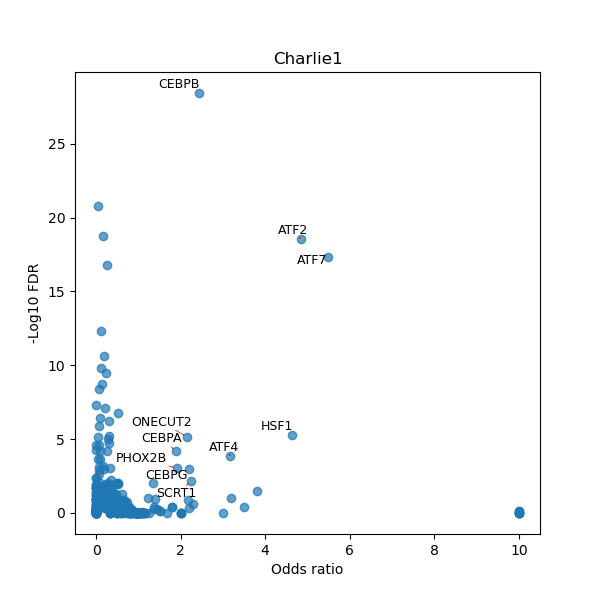
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.