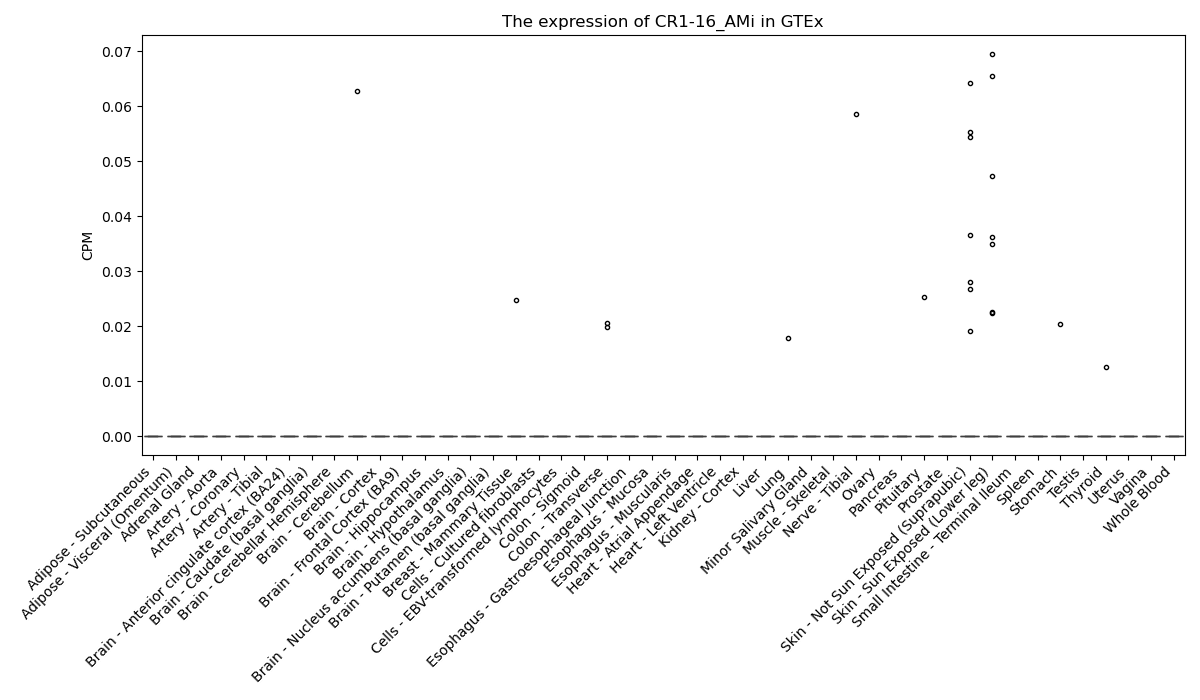
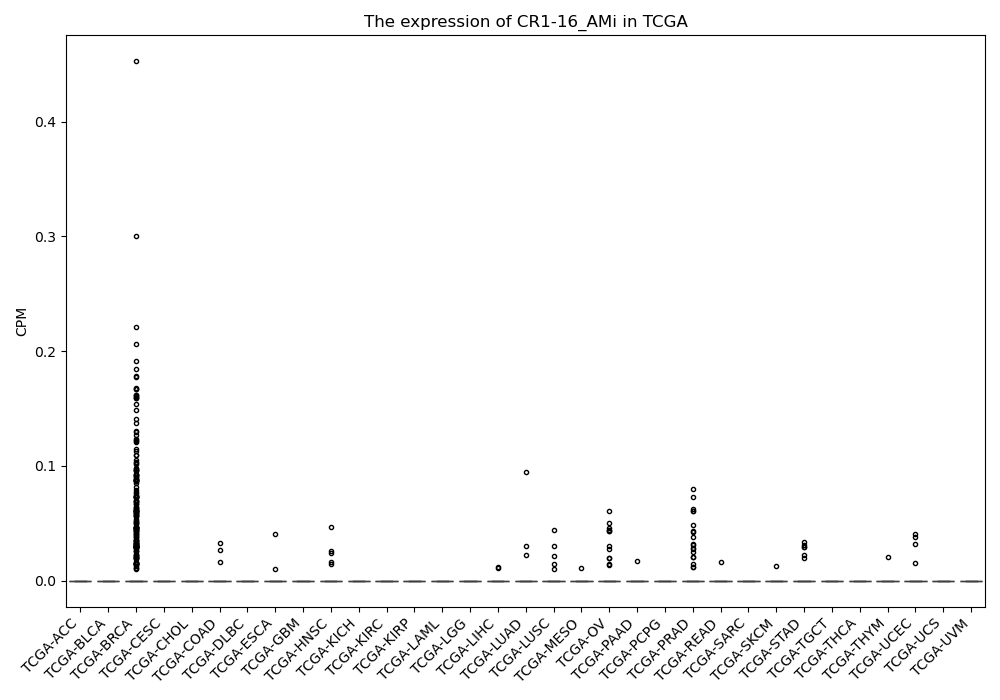
CR1-16_AMi
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0001265 |
|---|---|
| TE superfamily | CR1 |
| TE class | LINE |
| Species | Amniota |
| Length | 851 |
| Kimura value | 31.52 |
| Tau index | 0.0000 |
| Description | Non-LTR retrotransposon. |
| Comment | - |
| Sequence |
AACTTATTGTAACATGGAGCATTATTTTGAAAAGTCACGAAAANTGGGNAGGTACCGGAAGATTGGAGAAAAGCAAACGCGCTGATGTCTTCAAAAAAGGAAGGAGAGTGATCCTGGCAATTACCATCCTCAAGTCTTAACTTCTATCCCTAGTAAAATAATGGAACAAATATTAAGAGGGAGGAAAAAATCACTAAACATCTAGAGGATAACGGAACCATGAGCAATAAGCAGCATGGGTTTATAAAGAACAAATCTTGCCAGACAAACTTGATTGCTTTTTTTGATAGAATTACAAAATTAGTGGATGAAGGGATGCAATATATTTGGATTTTAGTAAAGCATTTGATACAGTGTCTCACATATCTTAAAATTAATTCAAATTGGCTTGGATATGAGCACTGTCACGTGGACTGAAAACTGGCTGAAGGACCGTAAACAAAGGGTAATGATAAACGGCAATGTATCGAGTTGGGGGGAGGTGTCTAGGGGTACTGCAGGGATGGTGTTAGGTCTGGTCTTATTTAACATCTTCATTAATGATCTGGAAGAGGGAGTAAACAGCACGTTAATGAAATTCGCAGATGATACTAAATTGGGAGGAGTTGCAAACACCAGTGAGGACAGAGAAATAATACAAAGGGACCTAGAGAGATTAGAAAGGAAATAACAAAATGAGATTAACTTGGAAAAATGCAGCTAATATTCACTGGGGAAAATCCAATGGAAGAGGAAACNTGGAAAGCAGTAATGANAGAGACCTNGAGGCGATAGCGGACAGCAAATTAGATATGAGTTGCAATGTGATATGGCAGCAAAAGGCCAGTGCAATTTTGGGCTGCATATGCGGA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| CR1-16_AMi | E2FD | 806 | 820 | + | 3.43 | GATATGGCAGCAAAA |
| CR1-16_AMi | E2FE | 806 | 820 | + | 3.43 | GATATGGCAGCAAAA |
| CR1-16_AMi | NFIC | 63 | 77 | + | 1.67 | TTGGAGAAAAGCAAA |
| CR1-16_AMi | ZNF143 | 302 | 317 | - | 1.55 | ATCCCTTCATCCACTA |
| CR1-16_AMi | BPC1 | 165 | 188 | + | 0.11 | AACAAATATTAAGAGGGAGGAAAA |
| CR1-16_AMi | EWSR1-FLI1 | 91 | 108 | + | -11.45 | TCAAAAAAGGAAGGAGAG |
| CR1-16_AMi | EWSR1-FLI1 | 306 | 323 | + | -15.07 | GGATGAAGGGATGCAATA |
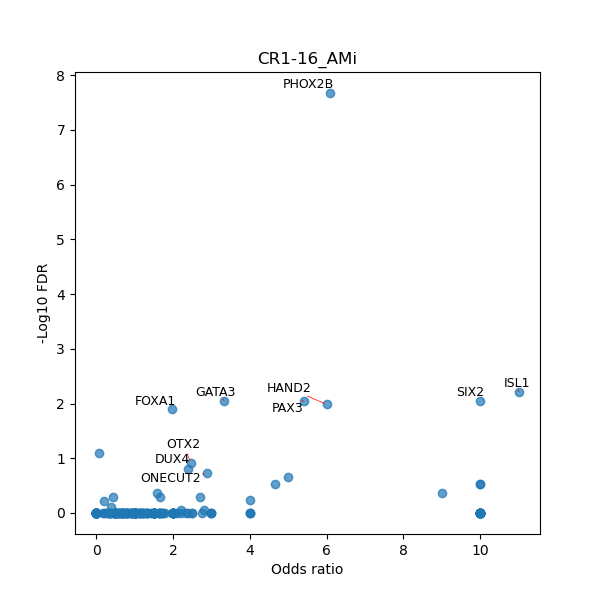
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.