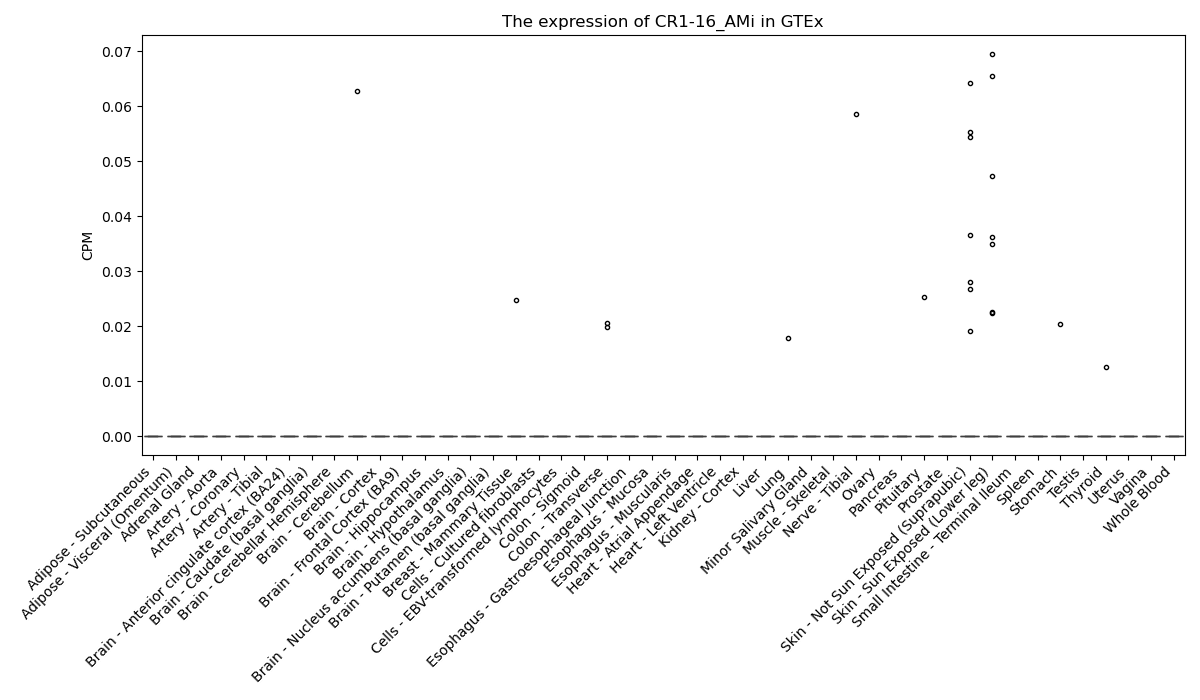
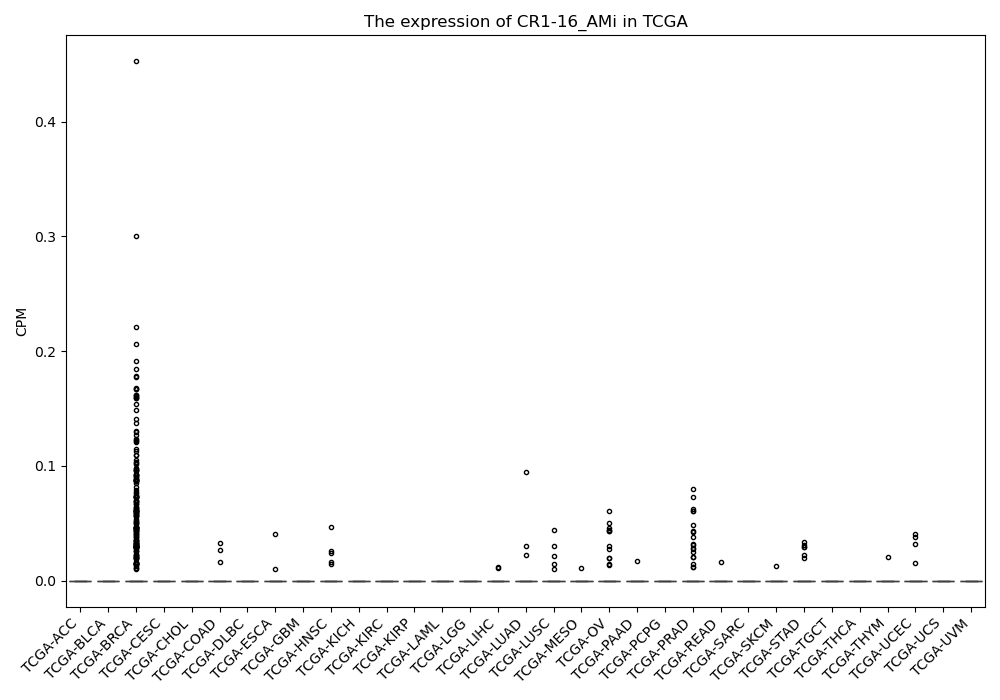
CR1-16_AMi
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0001265 |
|---|---|
| TE superfamily | CR1 |
| TE class | LINE |
| Species | Amniota |
| Length | 851 |
| Kimura value | 31.52 |
| Tau index | 0.0000 |
| Description | Non-LTR retrotransposon. |
| Comment | - |
| Sequence |
AACTTATTGTAACATGGAGCATTATTTTGAAAAGTCACGAAAANTGGGNAGGTACCGGAAGATTGGAGAAAAGCAAACGCGCTGATGTCTTCAAAAAAGGAAGGAGAGTGATCCTGGCAATTACCATCCTCAAGTCTTAACTTCTATCCCTAGTAAAATAATGGAACAAATATTAAGAGGGAGGAAAAAATCACTAAACATCTAGAGGATAACGGAACCATGAGCAATAAGCAGCATGGGTTTATAAAGAACAAATCTTGCCAGACAAACTTGATTGCTTTTTTTGATAGAATTACAAAATTAGTGGATGAAGGGATGCAATATATTTGGATTTTAGTAAAGCATTTGATACAGTGTCTCACATATCTTAAAATTAATTCAAATTGGCTTGGATATGAGCACTGTCACGTGGACTGAAAACTGGCTGAAGGACCGTAAACAAAGGGTAATGATAAACGGCAATGTATCGAGTTGGGGGGAGGTGTCTAGGGGTACTGCAGGGATGGTGTTAGGTCTGGTCTTATTTAACATCTTCATTAATGATCTGGAAGAGGGAGTAAACAGCACGTTAATGAAATTCGCAGATGATACTAAATTGGGAGGAGTTGCAAACACCAGTGAGGACAGAGAAATAATACAAAGGGACCTAGAGAGATTAGAAAGGAAATAACAAAATGAGATTAACTTGGAAAAATGCAGCTAATATTCACTGGGGAAAATCCAATGGAAGAGGAAACNTGGAAAGCAGTAATGANAGAGACCTNGAGGCGATAGCGGACAGCAAATTAGATATGAGTTGCAATGTGATATGGCAGCAAAAGGCCAGTGCAATTTTGGGCTGCATATGCGGA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| CR1-16_AMi | dm | 406 | 413 | - | 13.75 | TCCACGTG |
| CR1-16_AMi | E(spl)mgamma-HLH | 403 | 412 | - | 13.75 | CCACGTGACA |
| CR1-16_AMi | Max | 406 | 413 | - | 13.75 | TCCACGTG |
| CR1-16_AMi | PIF3 | 406 | 412 | - | 13.71 | CCACGTG |
| CR1-16_AMi | let-381 | 435 | 442 | + | 13.69 | GTAAACAA |
| CR1-16_AMi | FKH1 | 431 | 443 | + | 13.61 | GACCGTAAACAAA |
| CR1-16_AMi | TYE7 | 405 | 411 | - | 13.51 | CACGTGA |
| CR1-16_AMi | nuc-1 | 406 | 412 | - | 13.51 | CCACGTG |
| CR1-16_AMi | D1 | 154 | 161 | + | 13.51 | TAAAATAA |
| CR1-16_AMi | Ets97D | 54 | 62 | + | 13.46 | ACCGGAAGA |
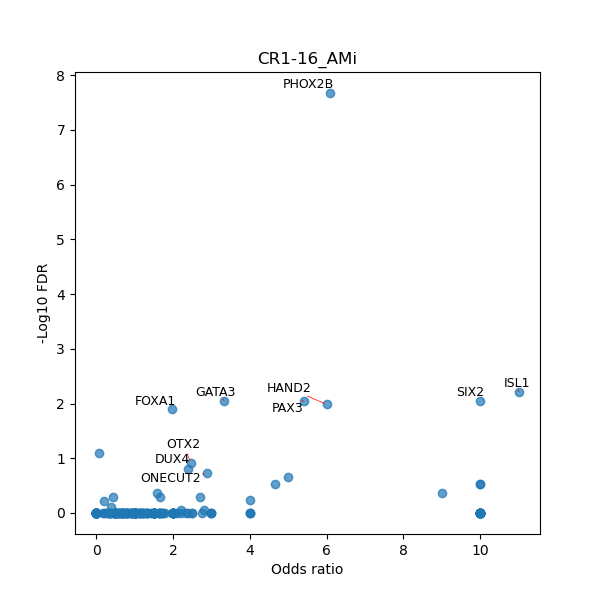
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.