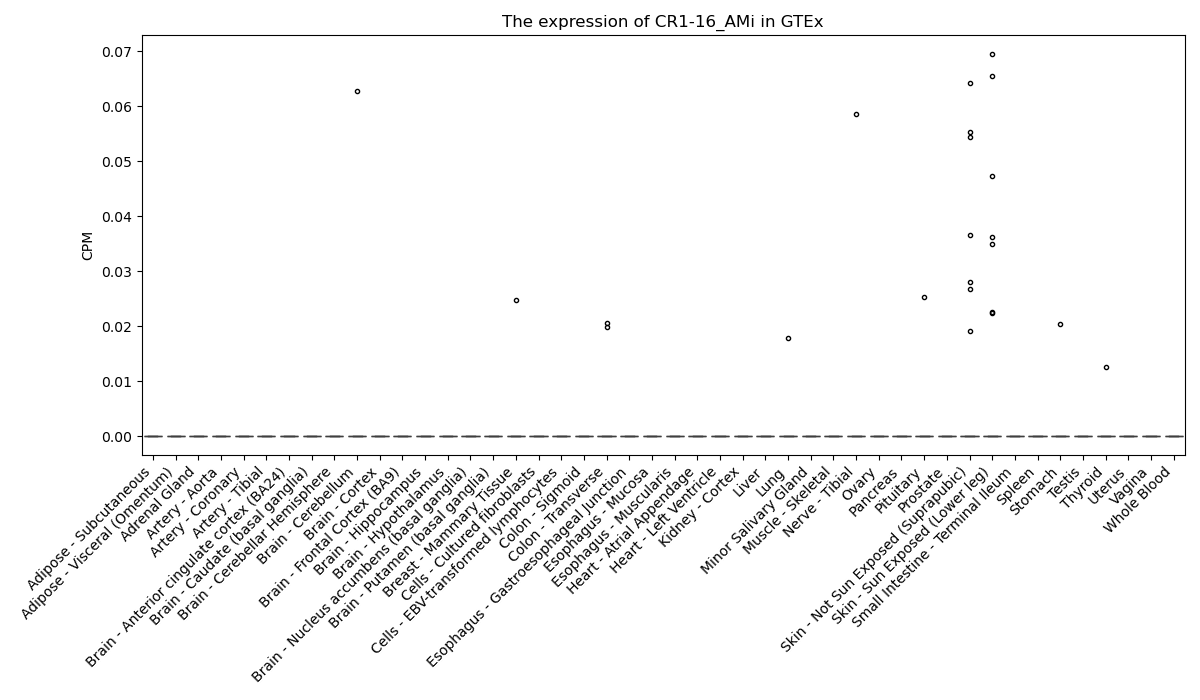
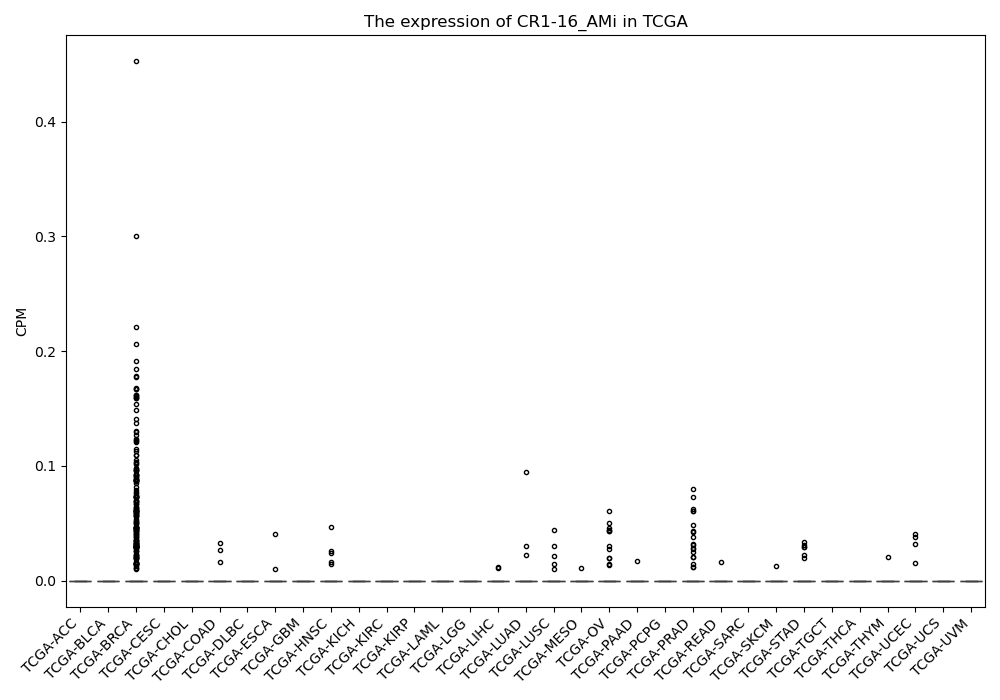
CR1-16_AMi
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0001265 |
|---|---|
| TE superfamily | CR1 |
| TE class | LINE |
| Species | Amniota |
| Length | 851 |
| Kimura value | 31.52 |
| Tau index | 0.0000 |
| Description | Non-LTR retrotransposon. |
| Comment | - |
| Sequence |
AACTTATTGTAACATGGAGCATTATTTTGAAAAGTCACGAAAANTGGGNAGGTACCGGAAGATTGGAGAAAAGCAAACGCGCTGATGTCTTCAAAAAAGGAAGGAGAGTGATCCTGGCAATTACCATCCTCAAGTCTTAACTTCTATCCCTAGTAAAATAATGGAACAAATATTAAGAGGGAGGAAAAAATCACTAAACATCTAGAGGATAACGGAACCATGAGCAATAAGCAGCATGGGTTTATAAAGAACAAATCTTGCCAGACAAACTTGATTGCTTTTTTTGATAGAATTACAAAATTAGTGGATGAAGGGATGCAATATATTTGGATTTTAGTAAAGCATTTGATACAGTGTCTCACATATCTTAAAATTAATTCAAATTGGCTTGGATATGAGCACTGTCACGTGGACTGAAAACTGGCTGAAGGACCGTAAACAAAGGGTAATGATAAACGGCAATGTATCGAGTTGGGGGGAGGTGTCTAGGGGTACTGCAGGGATGGTGTTAGGTCTGGTCTTATTTAACATCTTCATTAATGATCTGGAAGAGGGAGTAAACAGCACGTTAATGAAATTCGCAGATGATACTAAATTGGGAGGAGTTGCAAACACCAGTGAGGACAGAGAAATAATACAAAGGGACCTAGAGAGATTAGAAAGGAAATAACAAAATGAGATTAACTTGGAAAAATGCAGCTAATATTCACTGGGGAAAATCCAATGGAAGAGGAAACNTGGAAAGCAGTAATGANAGAGACCTNGAGGCGATAGCGGACAGCAAATTAGATATGAGTTGCAATGTGATATGGCAGCAAAAGGCCAGTGCAATTTTGGGCTGCATATGCGGA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| CR1-16_AMi | E(spl)m3-HLH | 403 | 411 | - | 14.05 | CACGTGACA |
| CR1-16_AMi | Fer2 | 403 | 412 | - | 14.05 | CCACGTGACA |
| CR1-16_AMi | TCX3 | 375 | 386 | + | 14.02 | TAATTCAAATTG |
| CR1-16_AMi | lin-54 | 373 | 385 | + | 13.90 | ATTAATTCAAATT |
| CR1-16_AMi | ZNF263 | 179 | 185 | + | 13.83 | GGGAGGA |
| CR1-16_AMi | ZNF263 | 598 | 604 | + | 13.83 | GGGAGGA |
| CR1-16_AMi | Hand1 | 511 | 519 | - | 13.82 | ACCAGACCT |
| CR1-16_AMi | MLXIPL | 404 | 411 | + | 13.82 | GTCACGTG |
| CR1-16_AMi | FOXG1 | 435 | 442 | + | 13.80 | GTAAACAA |
| CR1-16_AMi | PK05451.1 | 405 | 412 | - | 13.77 | CCACGTGA |
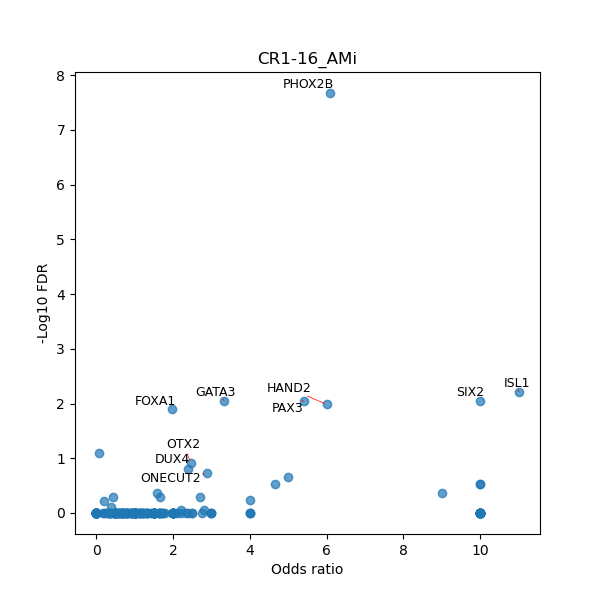
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.