Arthur1
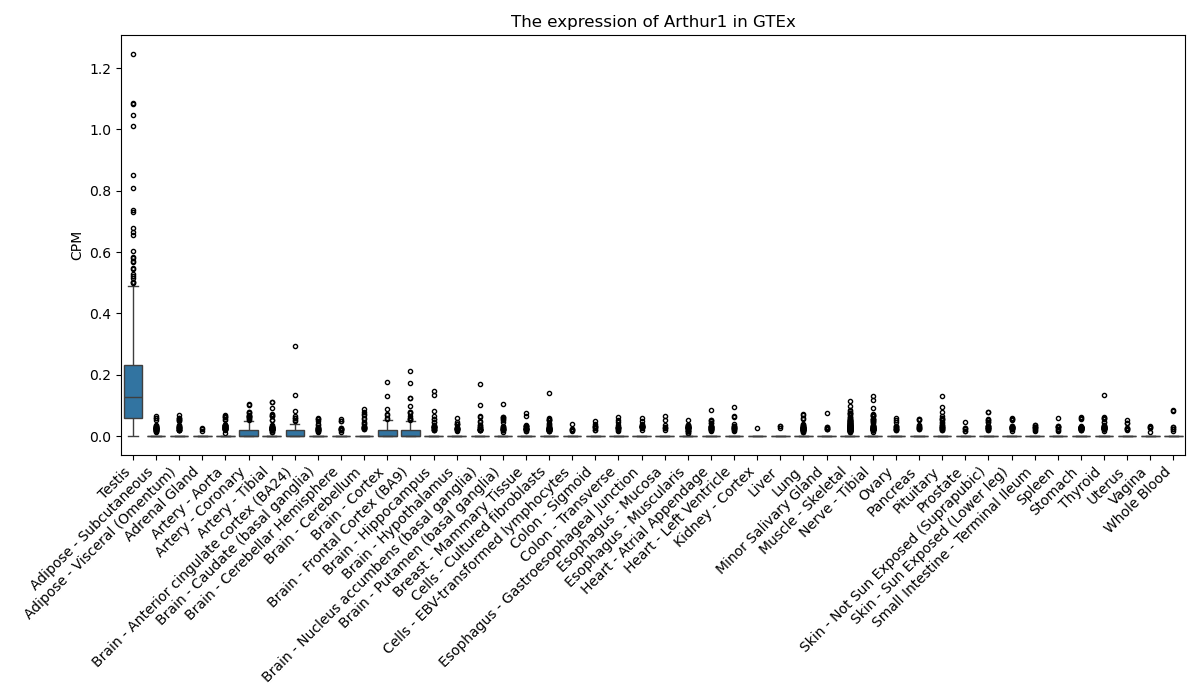
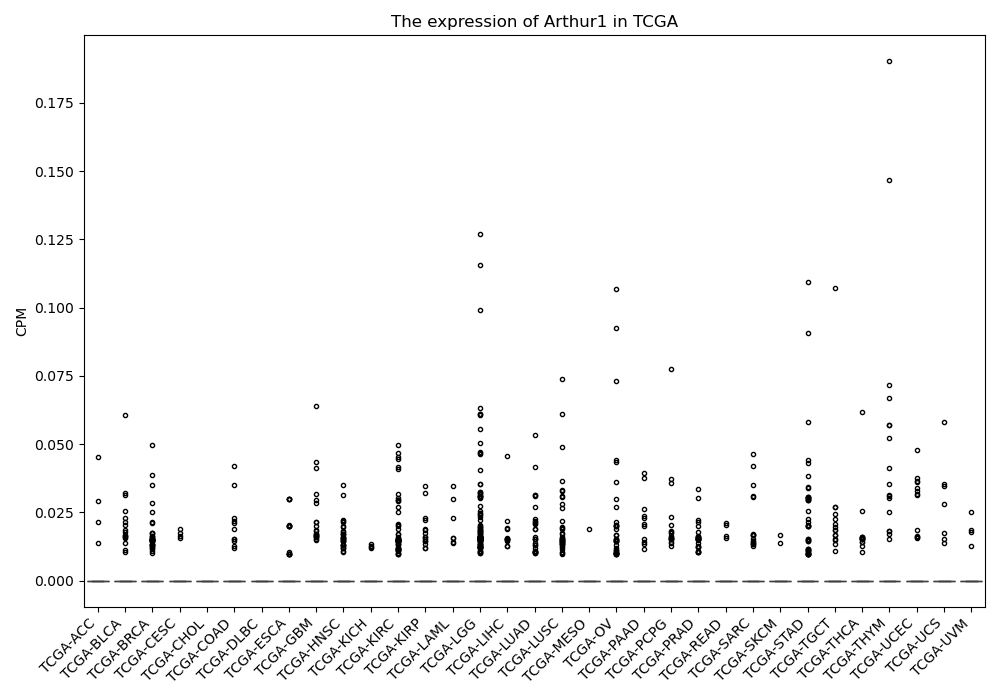
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000069 |
|---|---|
| TE superfamily | Tip100 |
| TE class | DNA |
| Species | Eutheria |
| Length | 3947 |
| Kimura value | 24.05 |
| Tau index | 1.0000 |
| Description | hAT-Tip100 DNA transposon, Arthur1 subfamily |
| Comment | Arthur1 has 8 bp TSDs and 11 bp TIRs. A complete ORF (~1150-3711) encodes a transposase related to that of the Tip100-group of DNA transposons. Note that MER69C is an incomplete reconstruction of an autonomous hAT-family DNA transposon related to MER45 and Zaphod. Arthur1 is a more complete reconstruction. |
| Sequence |
CAGAGGCGGATTTACCGTGAAGCTAATGAAGCTTAAGCTTCAGGGCCCCTCACTTGCACGGGCCCCTTCCAAGGCCCTGGGAGGGGCCCTAGCAATGTGTTCACATGGTCATATGTTTTTGTAAAATTTGCAAAAGTAAGATATTTTAACCGCAATCGGTTAAGACCGCTGTCTCTTTCCACTCCGACTTCCCCTCCGTCACACTTCCCCTCGTGTCGGGTGGCGTTGGAGTGGCCGCGGGCATTTTGGGGATCCGGCTAAGGGGAAGTTGAGTTGGGGATACATTTAGTTTGGGTTTAGTGGGATATATTTATGTGGTTCGCAGTCACTTCCGTGTATAGTTAAGTTATTGCTAGCCGTCCCGGTGTAGGAATGGCTTCCAGGAATACTCCTACCGCCCACTGTGCCGACTCACCCGGCGTCGTGACACGAAGGTGCAGGGCCAGAGGTCGTATCGCGATATGAACGTGTCCTACGGCGCCCGGCACCGGAAGTATGTGGGTAGTGGAGGAGAAACAAGGTTTGAAATGTACGGAGCCAGAAGCTAGTCTGTGGAAAATTCTTCCAATCATCAGACGTGTAAAATTGTAAGCGGAGGATTCGGTTCTCATCGACGCCTAGTCAAAACGGAAGTTCTCTCCTGTCAGGAATATACTCGATAATGCAGCATATACAATTATAAACGCACCATACATTTTTTTTCTTTTTTGATGGGAATCGCGCGAAATAGAATTTATCAGAATTCCTGTGTTTGTAGGGCACAAACCTGTAGCAGTACTACAAACAGCGAGTACGTCTGTGTGTGAAGTCGCATGTTTTATGCATCCCAACGTATCGGATCGCATCTTAGCGTATCTGACGCATCTTGTCCTGACGAAGTCTGGCGGTTCCAGACGAAACGGCACGCAAAATTGCCACCGACGCAGCGAAACGGCCTGAAGAAACAAGTGTTCCAGTGACAAAACTCCTACACATATTCATCAATAAGTCAATGTATGTAGTATAAGAGTAATTTGATTACACAATTATGTAGCTCAATACAACGTAGCGGCAATTTNAAAATGCATTTGAATTGCTCTTGTGTATGCCTTAATTTCAGATTAATTTTGTATATTTTTTCAGAATTATTGATCATACCAAAATTCAAATATGACGGGAAGGAAGTATCCTAGTGGAAGCCAAAAACGTAAACTGAAAGAAAGAAGACTGGCAGAAGCTTCAAAATGTCAAAAACTCTCTTATTATANCACAACGGAAACTAAAAACTCTGATGGTCAAAATCATGAAAATAAGGAAAAGAATCACGATAGCCAAGAAAATATAGATAATAAAAATGACAACAGAAGAGATAAAGAAGATTATGAGAATCAGGACACTCCCAAACAGGAAACAAAAAAGCAGAAGGAAGATGAAAATAATAGTAAAGAAAAACACAATTTGTATGGTCAACAGAGCACGCACGAAGGTCAGCAGCAGCTTCATTTCGACGAATTCGTTGACTTCGATAAGTACCGTGATCTTTCGTTATGNCCTGTTACAATGAGCAATAATTTCATTCAACACTGTTTAATNAAAGAAGTTTGTTTTTTCAAAATCGTGATCCCAATAATATTTACAGAGAATCAAACAGAACATACAATGGACAAAAGAGATGTTTCTCAAACAGATATTTTGAAAAAAATTAAAAATGGTCAAACTNTTATGAGATCCTGGCTAGCCTATTCCAAGACTAAAGGTTGCATATATTGTTTTGTTTGTAAACTGTTTTCAACAAGACAGACATTACTTANATCAGATGAGTTTTCAGACTGGATGAATATATTGAGAACCCTCAAAAGTCATGAAGACTCCACAGAGCACAAGAAGGCTATGTTTACTTGGATTACACGTAAATCAAATAAAAATGCACTAGACCAATGTCTTGAAGAACAGAGAAAGAACATTCAGTATTATTTTGAAGTGCTAAAAAGAGTAGTTGCGGTTATTAAATTTTTAAGCGAAAGAGGTTTGCCCTTCAGAGGTCACGATGAAAAGTGGCATTCTTCAAATAATGGAAATTTCATGGGAATCATCAAACTAATTGCTGAATTTGATCCATTTTTGCACGAGCACTTGGAAAAATNTCAAAATGAAAAAACAAATGTGACTTATTTATCCAAAACAGTTTATGAAGAATTAATCGAAATAATGGGAAAACGTGTACAAAATGAAGTGGTAAATCAAATAAATAAACCAGACACCAAATACTACTCCATTATTGTAGATTCTACACCGGATGTGACAAATACTGATCAGTTGGCGATTATTGTGCGCTACTGTTATAACGGAAAACCCTATGAGCGATTTTTAACTTTTTCGCCAACTGAAAACCATCTATCTGTGACTTTATTCAACAAAGTGAAACAAGTTCTGGACGATTGTAACCTGCCGTTGAACAACATTCGCGGTCAGTCATATGACAATACAGCTGTTATGAAAGGTGAGGACAAAGGGCTGCAAGCGCTCTTTAAAAACATTAATAAGTACGCAGAATATGTGCCGTGCGCAGCTCATTCCCTTAATCTCGTCGGAGAAAAAGCTGCCTCCACAGTACCTGAAGTTGTTGACTACTTCGGTATTTTGCAACAGTTATATGTTTTCTTTTCGGGTTCGTCTCGTAGATGGAGTATTTTAAACACACATGCCAACTTGGATTTTTCTTTAAAGAGTCTAAGTGTAACGAGATGGTCTGCCCATTATAAAGCTGTTCAAGCCTTGCAACATGGGTACAACGACATTTTAAGGACTCTAAAATATATTTTCGAAGACTCGGAAGAGAAACCCGAACACAAACGAGATGCAAAAAGTTTATTCAAAAAGTTGATAAAGCTCGAATACGCCATTTTAACTGCAATTTGGAAAGACGTTTTGGAACGTTTCAATAAAACAAGTGAGAAATTACAGACTCCTGATTTGGACGTATATGAAGGGTACCTTCTTTTGTCATCTTTAAATTTATTTATTAAAGAACTGAGAGAAAATTCAGATAAAAAATTAATGGAATATGAAACAAAAGCGATAAAGATGAGCGATGAAATCAATAGAAATTATTCTGACATCGAGAAGCAAATTGTAACAAAAAAATTTTCAGATCATACCAAAAGCAGTAATTCATTAAGAGGAAGAGATAAATTTCGAATAGAAGTAATGAATAGGCTCTTGGATTGTTTGATAATCCAATTAATGAAGAGGAGTGAATCATATGAACACATTGGAAAAAGATTTAAATTTCTTGCTGATTTGACACGAAATACTGACATCGACGAAGATAACATAAAACTCATAATACGCCACTACAACGAGGACATCGACGACAAACTGGTTAATGAGTGTCGTCAATTCAAAGAGTATTTAAGATTAGTCACTGCACAAGAAAACTTGAAATGCCCTGAAATCTTACAGCTCATATATGAAAGAAACTTGATAGAGGTTTTCCCAAATTTGACAACAATCCTAAAAATTTACATGACATTACCAATAACGAGTTGTGAAGCTGAAAGAAACTTTTCTAAACTATCAATAATAAAAAACAAATTTCGATCAACCATGCTAGAGGAAAGACTGAATTATCTTTCTATTCTCTCTATAGAAAATGATATTACAAAATCGTTGTCATATGAAGAGGCGATCAAAGAGTATGCAGCCAAAAAATGTAGGAAAAAAGTATTATAGAGGTGTGTCAGGCAGTTAATTAATAAAAATATTATGTTATTTTTCTGGATTTTGTGATGTTTGTGGTATTTGTCAGCTTTTTAAAATTTGTAATTTGTTGTGATTTCTTTTCTCATTCTAAATAAATATTCACTTTCGTACCTAATTTTGTATTCGTAATTTTGTATTCTTTTTCTTAAAGAGGGCCCCCCAAATTGTATAAGCTTCAGGCCCCACAAAACCTGGATCCGCCCCTG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Arthur1 | BPC5 | 180 | 209 | - | -46.12 | GGGAAGTGTGACGGAGGGGAAGTCGGAGTG |
| Arthur1 | BPC5 | 176 | 205 | - | -47.43 | AGTGTGACGGAGGGGAAGTCGGAGTGGAAA |
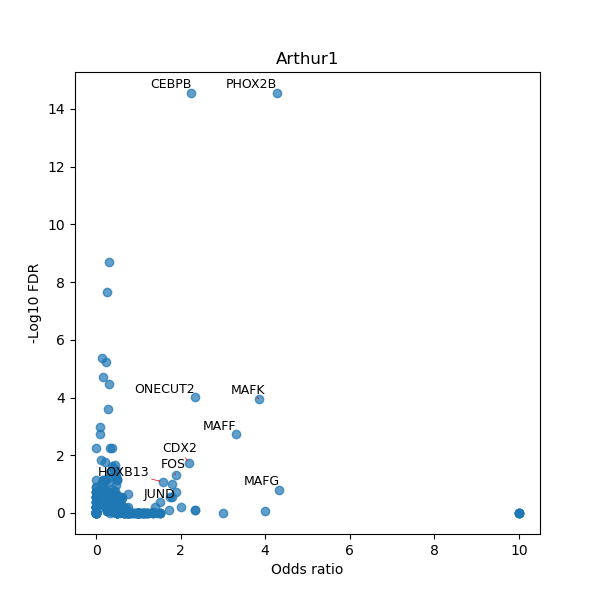
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.