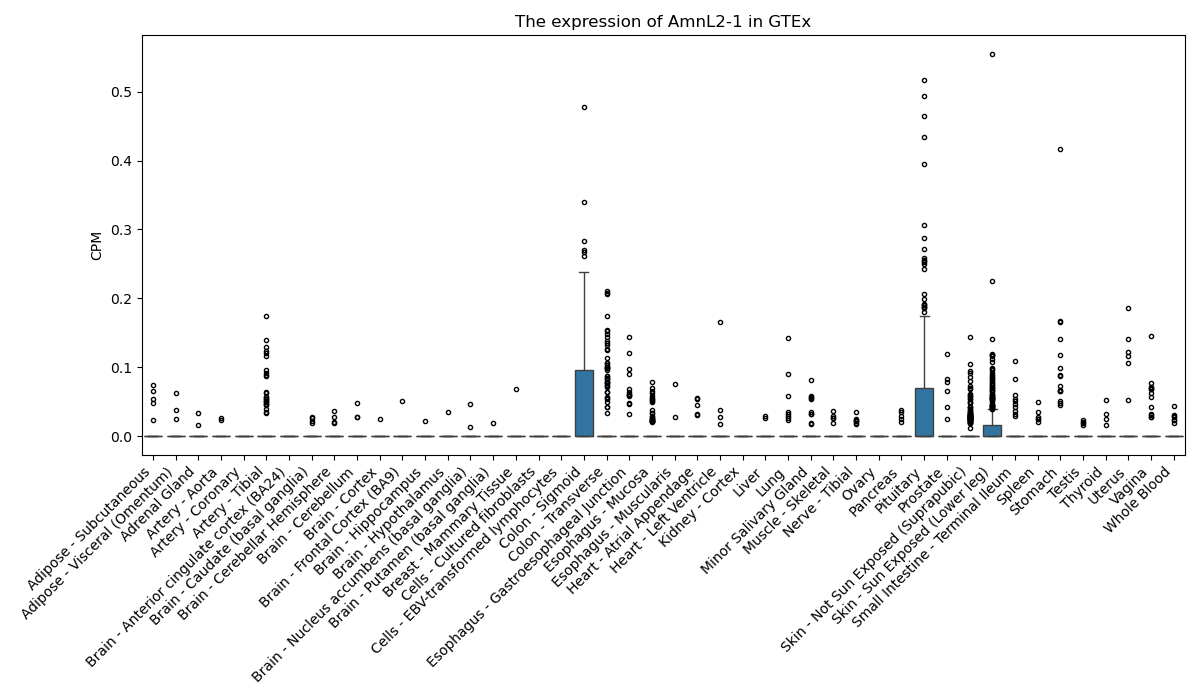
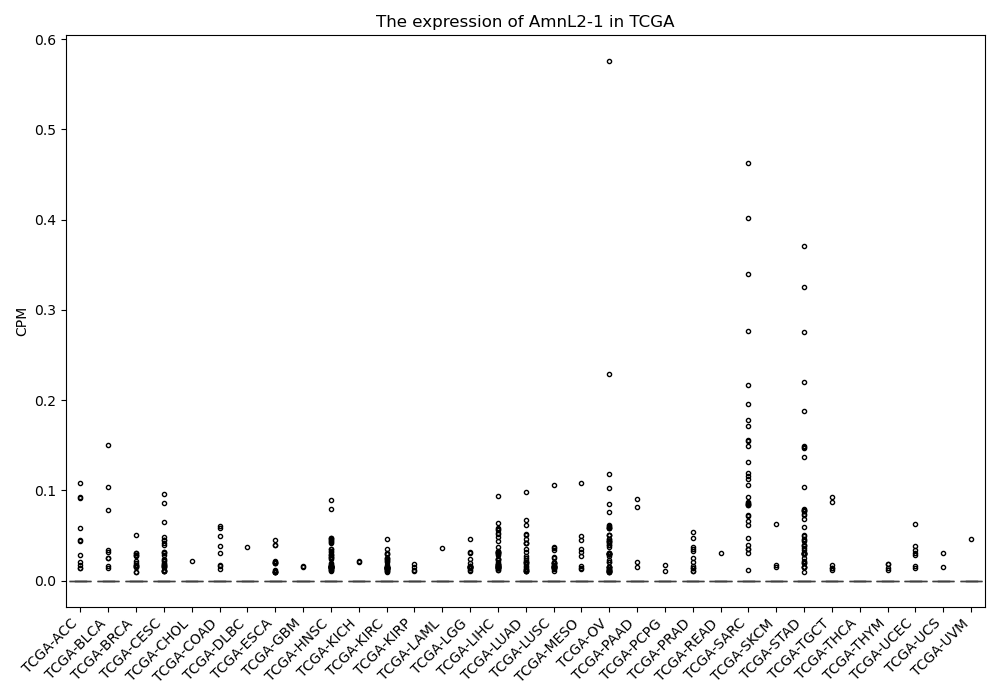
AmnL2-1
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0001732 |
|---|---|
| TE superfamily | L2 |
| TE class | LINE |
| Species | Amniota |
| Length | 2613 |
| Kimura value | 33.15 |
| Tau index | 0.0000 |
| Description | AmnL2-1 subfamily |
| Comment | The ORF 2-2488 encodes (part of) a pol most closely (45% id., 63% sim.) related to CR1-8_DR_pol and CR1-9_DR_pol in zebrafish. Pos 2396-2533 correspond to UCON49. Currently, AmnL2-1a only differs by a 90 bp replacement of a 22 bp sequence at 2509-2532, but no sites seem to cosegregate. |
| Sequence |
CTTCATTCAAGTTCTCCATTTCCCCCTCCAATCTCCTCCCTTACTTCAAACATCTTATCCCTATGACTTCCTCTCCCCTACTGGTTTCTCCTTCTCTGATTTCATCCTCACTGGGAACTTCACTGCGCNTGTTGACGTCCCCCTTGACCTNCTTNNCTCCAACGTCCTCTTCATCCTTGNCTNCCATAGNCTTACCCANTCTATCATCTCTCNCACCCGTATCNACGGCCATGCCCTCGTCATCAGNAACGCTCNCCCTCCCCTTTCCTATTCCGTTCATGATCTTNCCTACGCNGGCCACTCCNTGGTTATGTTTGNGCACTCTTTTCACCGNCTCACCCCTCAGAAAATTACCCTATCTCACTGCTCTCTCAAGCACCTTGATCCCGCTAGACTCTCCAGCCTGATTTGCTCTCTCGATCCCTCCTTCGCTGACCCTTCCTCCTCAGCCCCTTGCGTCGATGCCTANAACCCTGTCCTGTCTACTGNCCTTAACCAGCTCGCNTCCATCNANACNCGTACTGTTTCCTNCGCGCCAGATTCCCCCCCGCNTNCNGATCAGNTACGGAACTTCAAGTGTACAAACCGGCNCCTCGAAAAACAACGGCATATGTCCCATCTTCATGTCCGTTACGAGATCTGGAAATCTCATCAGATACTCTACAGGGATGCCCTCTGGGCTGCAAAGTCTAATTTCTACTCCTCCCTCACTTCCAAAGGTGCTGGTAATCCTTGCTCCCTCTTCCATACTATTAATTCTCNTCTCCACCCACCCAGCTCGCCCTTCCCTCCTAGCCTTTCCCCGGCCAGGTGCGAGGAATCTGCTTCCTTCTTTCAATCTAGGATTGCCCTAATTTGTGACANTATCTTCTCCNGCCCTANCACCTCCCTCCACTGATCCTTTCTCACTCCCCTTCTTCNCCTCTCCCTNTTNTCCCTGGACATTCCTGACATCATCCAGTCCCTNAAGAAAACTACCTCCTCTCTCGACCCCCTTCCNTCTCCTGTCGTTTGGTCCTGTCTCCCCACCCTTGCTCCCTACATTGCTACCACTGTCAATAACTCCTTTGCCTCTGGTACCTTCCCCAGCTCTCTAAAATCTGCCACTGTCCGTTCCCTTCTTAAAAAACCCTCCCTTGACCCCGACACTCTCTCTAACTACCGCCCTATCTCTAACCTTCCCTTTCTCAGCAAAGTGCTTGAGAAAGTAGCCTTCTCTCATCTCTCCAGTCACCTCTCACACAACANTATCCTTGACCGNTTTCAGTCTGGGTTTCGCTCTGGCCATAGCACGGAAACCGCTCTCCTCCGTGTAGTTAACGACCTCCTCCTTTCTGCTGATGANGGCTCTTTCTCCTTCCTCATTCTCCTTGACCTCAGTGCAGCCTTCGACACTGTGGACCATTCCATCCTCCTGGACCGCCTCCANAGCTATGCAGGCGTCTCTGGTTCCCTCCTGTCATGGTTCCGCTCCTACCTATCAAACCGCTCTTTCTCTGTCTCTGGTGGTAGCTCCTTCTCTGCGCCAAGACCCCTCCTCTATGGAGTCCCACAAGGTTCAGTTCTCGGCCCTCTGCTCTTTTCTATCTACCTTCTCCCTCTCGGTCATCTTNTCTCTACCTTTAACCTTAGCTACCACTTTTACGCTGATGATACTCAGCTGTACATATCTTTCGACACCTTTAATGACTCCTCCCTGTCCACCCTCTCTGCCTGTCTTGACCAAGTAAACTTGTGGCTTCGTCTCAGCTTCCTTCAGCTAAATGTCTCAAAGACAGAAGTGCTTCTCTTGGGCAAGAGGAGCATCCTCAAACACAACTCCATCTCCTCTATCTCCCTCCTCGACTCAGACCTCCCATTTGCCTCCGCTGTCCGTAACCTTGGCGTCATCCTTGACCCTGAACTCTCCTTCTCCTCCCACATCTCCTGTGTCTGCAAATCTGCCTATCACCACCTCCGCAACATTGCCCGTGTATGCCACTGCCTCGACTCCACCTCTGCTGAAATCCTCATCCATGCCTTCATCTCCTCTCGCCTTGACTATTGCAACTCCCTTCTCTATGGCCTCCCCAAGTCCCAGCTCTCCAGACTCCAGCATGTGCAGAATGCTGCTGCCCGCCTTCTCACACGTACAAAGAAGCGCGACCACATCACCCCCATCCTCAAACAACTCCACTGGCTCCCCATTCATTTCAGGATCCAGTTCAAAATTGCCTTCCTTGTTTTTAAAACCCTCCATGGACTTGCTCCAGCCTACCTCACGGACCTTGTCTCTCTCTACTCCCCTCCCCGCTCCCTCCGCTCGTCTAGCACCGGCCTCCTCTCTGTCCCTTCACACAATCTCGCCACTGTTGGTGCGAGAGCCTTCTCCTCTGCAGCTCCTGCCATCTGGAACAGCCTTCCTGTATCTCTCCGCCTCATCGACTCTCCATCTCNTTTCAAATCTCANCTGAAAACATATCTTTTCTCCCTTGCCTATACCTCTTAATTTCTCTCTCCCTCTGCTATTATTTATTATTTAACTCTGTAACTGTATTATTTAACTCTGTAAAGCGTTTTGGGATACAGTTTGTATGAAAGACGCTATACAAAATAAAGTTGTATTGTATTGTA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| AmnL2-1 | kni | 847 | 857 | - | 15.39 | AAATTAGGGCA |
| AmnL2-1 | mab-3 | 1954 | 1966 | - | 15.33 | AATGTTGCGGAGG |
| AmnL2-1 | ZNF549 | 2110 | 2117 | + | 15.24 | TGCTGCCC |
| AmnL2-1 | GRF4 | 2108 | 2115 | - | 15.23 | GCAGCAGC |
| AmnL2-1 | NHP10 | 800 | 807 | - | 15.18 | GCCGGGGA |
| AmnL2-1 | NAC094 | 1307 | 1322 | + | 14.95 | CTCCGTGTAGTTAACG |
| AmnL2-1 | TRB2 | 846 | 856 | + | 14.91 | TTGCCCTAATT |
| AmnL2-1 | SPIB | 64 | 76 | + | 14.85 | TGACTTCCTCTCC |
| AmnL2-1 | TFAP2C | 670 | 682 | - | 14.81 | AGCCCAGAGGGCA |
| AmnL2-1 | NEUROG1 | 606 | 615 | + | 14.76 | GGCATATGTC |
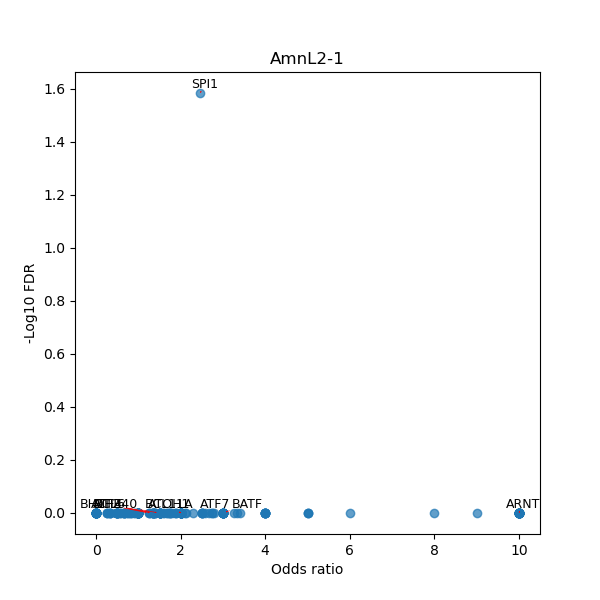
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.