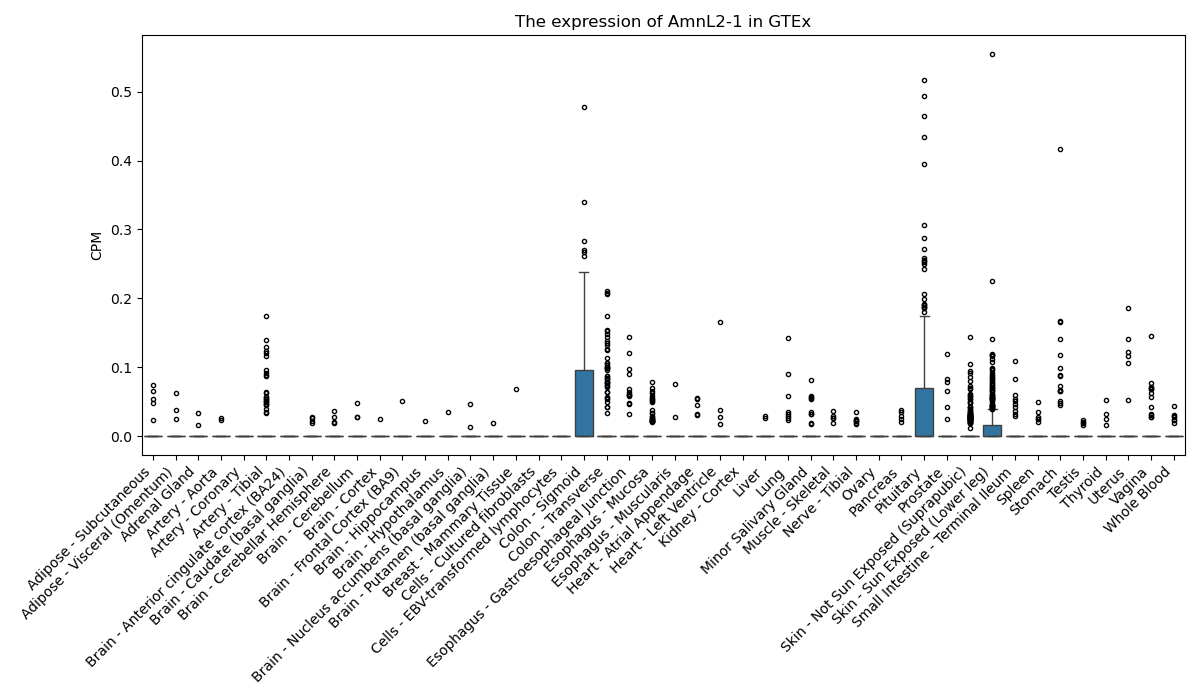
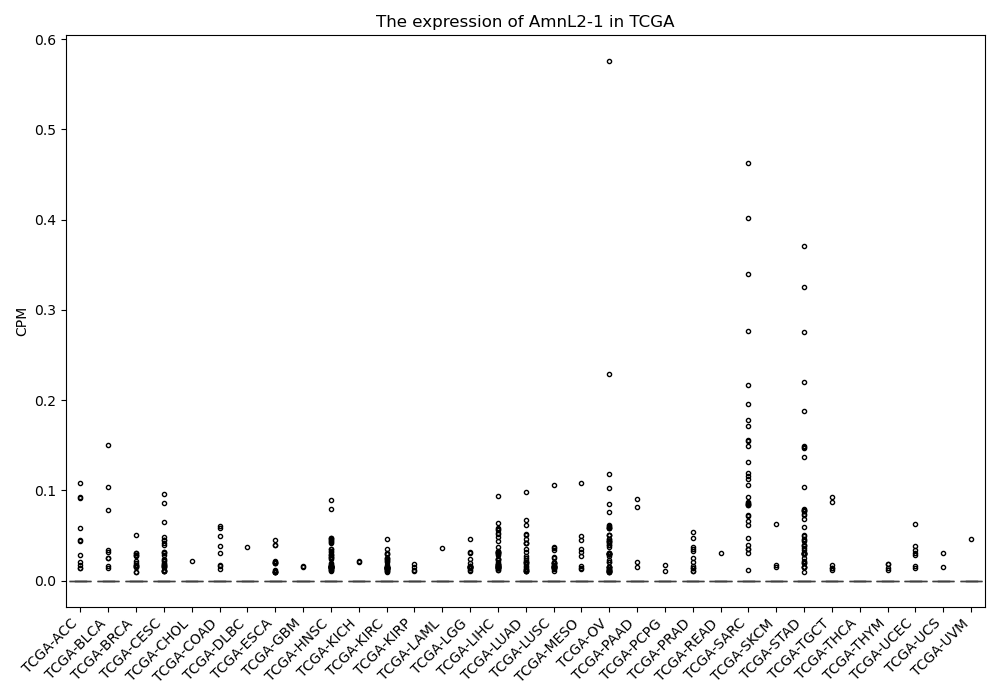
AmnL2-1
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0001732 |
|---|---|
| TE superfamily | L2 |
| TE class | LINE |
| Species | Amniota |
| Length | 2613 |
| Kimura value | 33.15 |
| Tau index | 0.0000 |
| Description | AmnL2-1 subfamily |
| Comment | The ORF 2-2488 encodes (part of) a pol most closely (45% id., 63% sim.) related to CR1-8_DR_pol and CR1-9_DR_pol in zebrafish. Pos 2396-2533 correspond to UCON49. Currently, AmnL2-1a only differs by a 90 bp replacement of a 22 bp sequence at 2509-2532, but no sites seem to cosegregate. |
| Sequence |
CTTCATTCAAGTTCTCCATTTCCCCCTCCAATCTCCTCCCTTACTTCAAACATCTTATCCCTATGACTTCCTCTCCCCTACTGGTTTCTCCTTCTCTGATTTCATCCTCACTGGGAACTTCACTGCGCNTGTTGACGTCCCCCTTGACCTNCTTNNCTCCAACGTCCTCTTCATCCTTGNCTNCCATAGNCTTACCCANTCTATCATCTCTCNCACCCGTATCNACGGCCATGCCCTCGTCATCAGNAACGCTCNCCCTCCCCTTTCCTATTCCGTTCATGATCTTNCCTACGCNGGCCACTCCNTGGTTATGTTTGNGCACTCTTTTCACCGNCTCACCCCTCAGAAAATTACCCTATCTCACTGCTCTCTCAAGCACCTTGATCCCGCTAGACTCTCCAGCCTGATTTGCTCTCTCGATCCCTCCTTCGCTGACCCTTCCTCCTCAGCCCCTTGCGTCGATGCCTANAACCCTGTCCTGTCTACTGNCCTTAACCAGCTCGCNTCCATCNANACNCGTACTGTTTCCTNCGCGCCAGATTCCCCCCCGCNTNCNGATCAGNTACGGAACTTCAAGTGTACAAACCGGCNCCTCGAAAAACAACGGCATATGTCCCATCTTCATGTCCGTTACGAGATCTGGAAATCTCATCAGATACTCTACAGGGATGCCCTCTGGGCTGCAAAGTCTAATTTCTACTCCTCCCTCACTTCCAAAGGTGCTGGTAATCCTTGCTCCCTCTTCCATACTATTAATTCTCNTCTCCACCCACCCAGCTCGCCCTTCCCTCCTAGCCTTTCCCCGGCCAGGTGCGAGGAATCTGCTTCCTTCTTTCAATCTAGGATTGCCCTAATTTGTGACANTATCTTCTCCNGCCCTANCACCTCCCTCCACTGATCCTTTCTCACTCCCCTTCTTCNCCTCTCCCTNTTNTCCCTGGACATTCCTGACATCATCCAGTCCCTNAAGAAAACTACCTCCTCTCTCGACCCCCTTCCNTCTCCTGTCGTTTGGTCCTGTCTCCCCACCCTTGCTCCCTACATTGCTACCACTGTCAATAACTCCTTTGCCTCTGGTACCTTCCCCAGCTCTCTAAAATCTGCCACTGTCCGTTCCCTTCTTAAAAAACCCTCCCTTGACCCCGACACTCTCTCTAACTACCGCCCTATCTCTAACCTTCCCTTTCTCAGCAAAGTGCTTGAGAAAGTAGCCTTCTCTCATCTCTCCAGTCACCTCTCACACAACANTATCCTTGACCGNTTTCAGTCTGGGTTTCGCTCTGGCCATAGCACGGAAACCGCTCTCCTCCGTGTAGTTAACGACCTCCTCCTTTCTGCTGATGANGGCTCTTTCTCCTTCCTCATTCTCCTTGACCTCAGTGCAGCCTTCGACACTGTGGACCATTCCATCCTCCTGGACCGCCTCCANAGCTATGCAGGCGTCTCTGGTTCCCTCCTGTCATGGTTCCGCTCCTACCTATCAAACCGCTCTTTCTCTGTCTCTGGTGGTAGCTCCTTCTCTGCGCCAAGACCCCTCCTCTATGGAGTCCCACAAGGTTCAGTTCTCGGCCCTCTGCTCTTTTCTATCTACCTTCTCCCTCTCGGTCATCTTNTCTCTACCTTTAACCTTAGCTACCACTTTTACGCTGATGATACTCAGCTGTACATATCTTTCGACACCTTTAATGACTCCTCCCTGTCCACCCTCTCTGCCTGTCTTGACCAAGTAAACTTGTGGCTTCGTCTCAGCTTCCTTCAGCTAAATGTCTCAAAGACAGAAGTGCTTCTCTTGGGCAAGAGGAGCATCCTCAAACACAACTCCATCTCCTCTATCTCCCTCCTCGACTCAGACCTCCCATTTGCCTCCGCTGTCCGTAACCTTGGCGTCATCCTTGACCCTGAACTCTCCTTCTCCTCCCACATCTCCTGTGTCTGCAAATCTGCCTATCACCACCTCCGCAACATTGCCCGTGTATGCCACTGCCTCGACTCCACCTCTGCTGAAATCCTCATCCATGCCTTCATCTCCTCTCGCCTTGACTATTGCAACTCCCTTCTCTATGGCCTCCCCAAGTCCCAGCTCTCCAGACTCCAGCATGTGCAGAATGCTGCTGCCCGCCTTCTCACACGTACAAAGAAGCGCGACCACATCACCCCCATCCTCAAACAACTCCACTGGCTCCCCATTCATTTCAGGATCCAGTTCAAAATTGCCTTCCTTGTTTTTAAAACCCTCCATGGACTTGCTCCAGCCTACCTCACGGACCTTGTCTCTCTCTACTCCCCTCCCCGCTCCCTCCGCTCGTCTAGCACCGGCCTCCTCTCTGTCCCTTCACACAATCTCGCCACTGTTGGTGCGAGAGCCTTCTCCTCTGCAGCTCCTGCCATCTGGAACAGCCTTCCTGTATCTCTCCGCCTCATCGACTCTCCATCTCNTTTCAAATCTCANCTGAAAACATATCTTTTCTCCCTTGCCTATACCTCTTAATTTCTCTCTCCCTCTGCTATTATTTATTATTTAACTCTGTAACTGTATTATTTAACTCTGTAAAGCGTTTTGGGATACAGTTTGTATGAAAGACGCTATACAAAATAAAGTTGTATTGTATTGTA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| AmnL2-1 | ZNF768 | 1704 | 1712 | + | 16.08 | CCCTCTCTG |
| AmnL2-1 | SP1 | 2283 | 2291 | - | 16.06 | GGGGAGGGG |
| AmnL2-1 | SP4 | 2283 | 2291 | - | 15.85 | GGGGAGGGG |
| AmnL2-1 | ZKSCAN5 | 707 | 715 | - | 15.83 | GGAAGTGAG |
| AmnL2-1 | ZNF324 | 1175 | 1188 | - | 15.63 | AGAAAGGGAAGGTT |
| AmnL2-1 | NR5A1 | 1894 | 1905 | - | 15.52 | AGTTCAGGGTCA |
| AmnL2-1 | Prdm5 | 11 | 21 | + | 15.52 | GTTCTCCATTT |
| AmnL2-1 | TFAP2A | 670 | 682 | + | 15.51 | TGCCCTCTGGGCT |
| AmnL2-1 | KLF4 | 1024 | 1031 | + | 15.48 | CCCCACCC |
| AmnL2-1 | NEUROG2 | 606 | 615 | - | 15.41 | GACATATGCC |
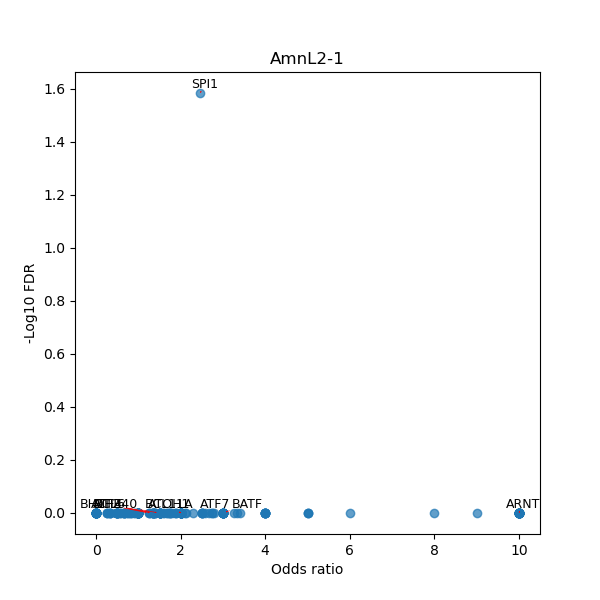
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.