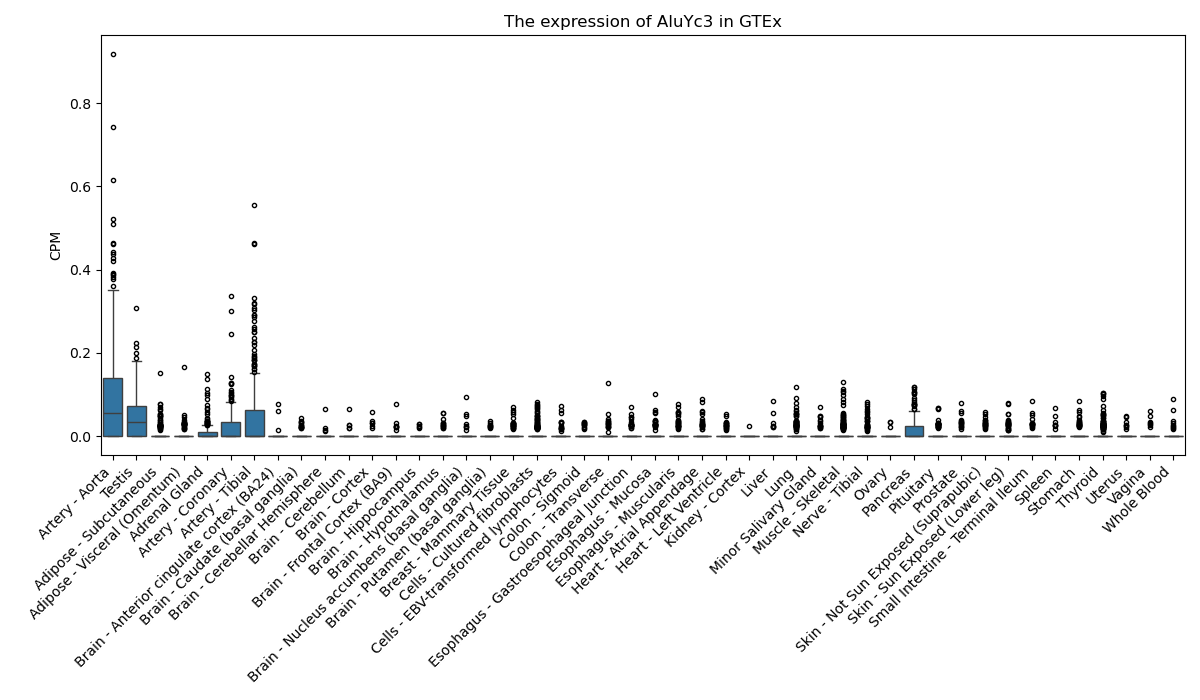
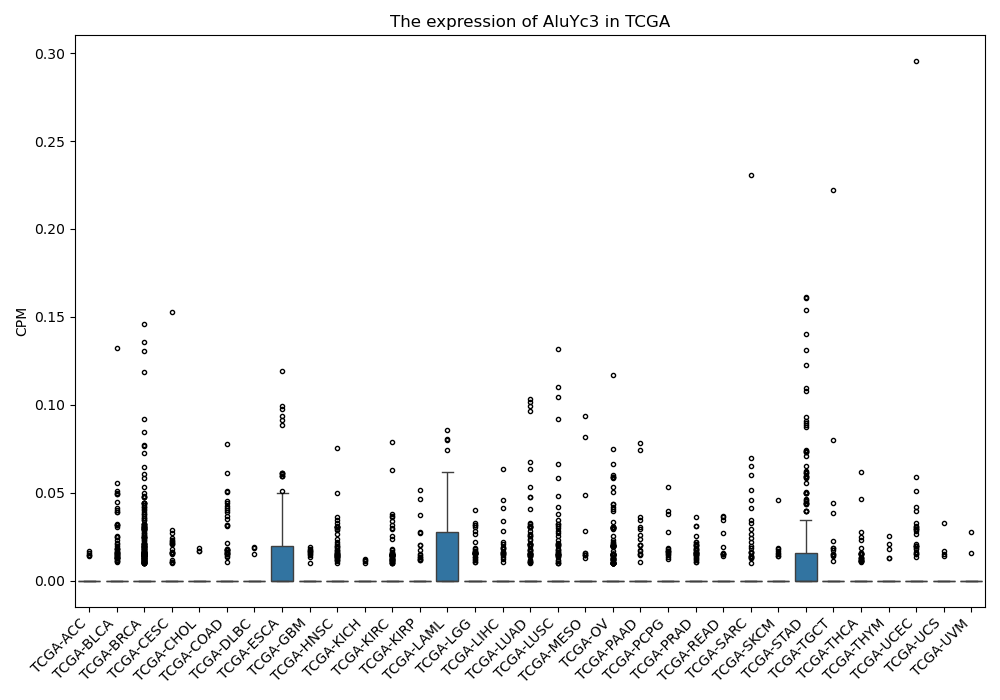
AluYc3
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000058 |
|---|---|
| TE superfamily | Alu |
| TE class | SINE |
| Species | Hominoidea |
| Length | 300 |
| Kimura value | 3.98 |
| Tau index | 0.9864 |
| Description | AluYc3 subfamily |
| Comment | Alu elements derive from a pair of internally-deleted 7SL RNA monomers that are joined by an A-rich linker. AluYc2, AluYc3, and AluYd3a1 are identical. Naming convention suggests AluYc2 is preferred, as seq differs from AluY at 2 positions. |
| Sequence |
GGCCGGGCGCGGTGGCTCACGCTTGTAATCCCAGCACTTTGGGAGGCCGAGGCGGGCGGATCACGAGGTCAGGAGATCGAGACCACGGTGAAACCCCGTCTCTACTAAAAATACAAAAAATTAGCCGGGCGTGGTGGCGGGCGCCTGTAGTCCCAGCTACTCGGAGAGGCTGAGGCAGGAGAATGGCGTGAACCCGGGAGGCGGAGCTTGCAGTGAGCCGAGATCGCGCCACTGCACTCCAGCCTGGGCGACAGAGCGAGACTCCGTCTCAAAAAAAAAAAAAAAAAAAAAAAAAAAAAA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| AluYc3 | luna | 127 | 135 | - | 15.26 | ACCACGCCC |
| AluYc3 | Klf3/8/12 | 127 | 135 | - | 15.24 | ACCACGCCC |
| AluYc3 | ZNF135 | 69 | 82 | - | 14.93 | TCTCGATCTCCTGA |
| AluYc3 | KLF3 | 127 | 136 | - | 14.83 | CACCACGCCC |
| AluYc3 | Spps | 197 | 207 | - | 14.75 | GCTCCGCCTCC |
| AluYc3 | ZNF257 | 172 | 181 | + | 14.53 | GAGGCAGGAG |
| AluYc3 | KLF3 | 5 | 14 | - | 14.43 | CACCGCGCCC |
| AluYc3 | CRF4 | 131 | 140 | - | 14.36 | CCGCCACCAC |
| AluYc3 | KLF2 | 127 | 134 | - | 14.29 | CCACGCCC |
| AluYc3 | EREB71 | 5 | 14 | - | 14.22 | CACCGCGCCC |
TFBS enrichment in GRCh38
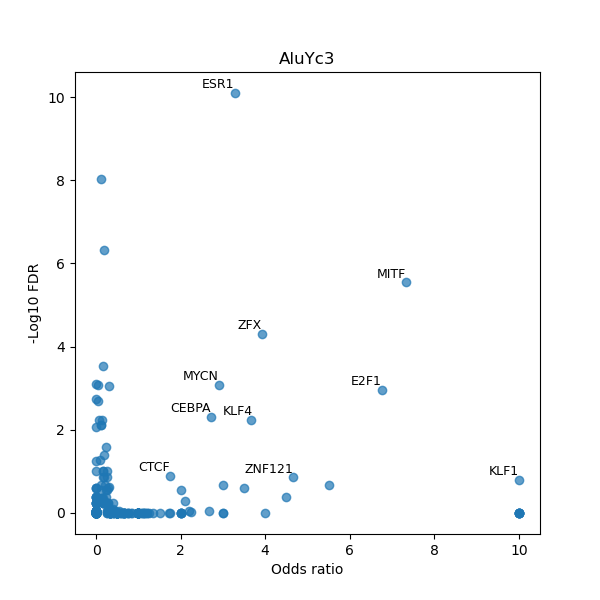
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.