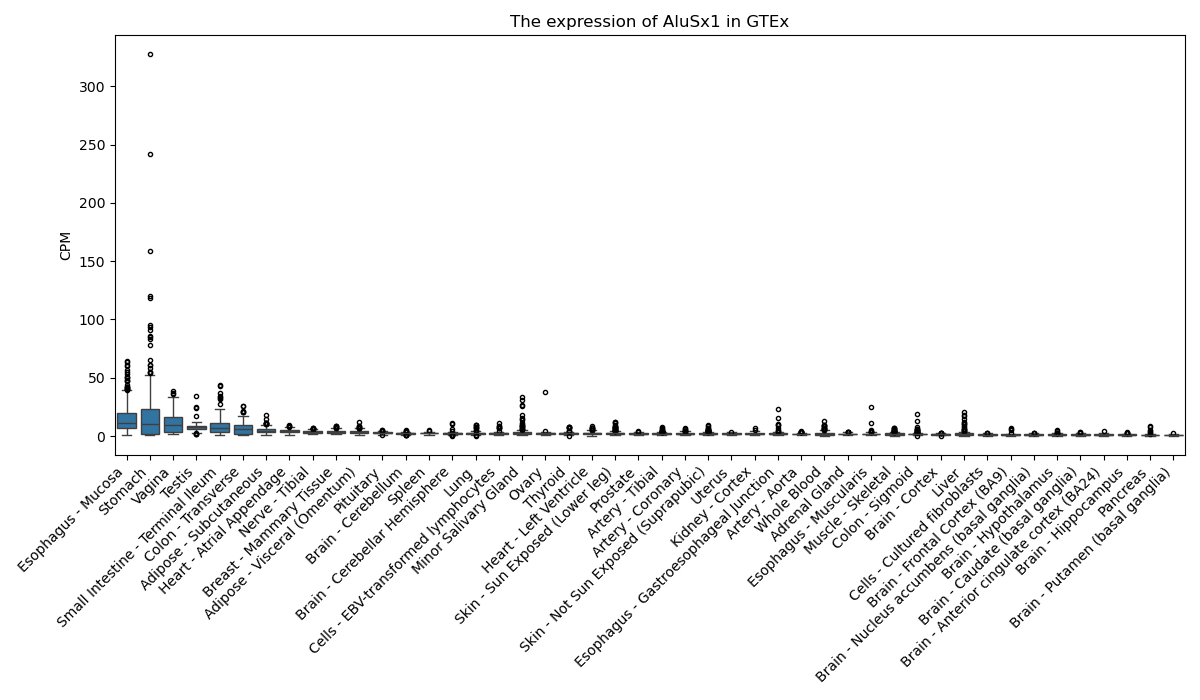
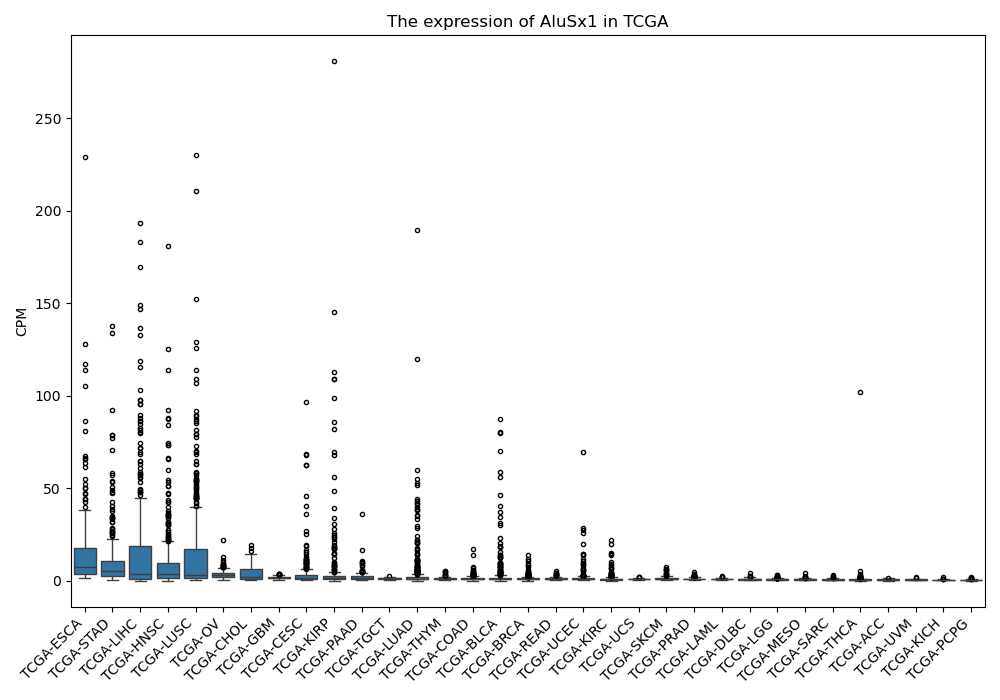
AluSx1
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000048 |
|---|---|
| TE superfamily | Alu |
| TE class | SINE |
| Species | Haplorrhini |
| Length | 312 |
| Kimura value | 7.10 |
| Tau index | 0.7579 |
| Description | AluSx1 subfamily |
| Comment | Alu elements derive from a pair of internally-deleted 7SL RNA monomers that are joined by an A-rich linker. |
| Sequence |
GGCCGGGCGCGGTGGCTCACGCCTGTAATCCCAGCACTTTGGGAGGCCGAGGCGGGCGGATCACCTGAGGTCAGGAGTTCGAGACCAGCCTGGCCAACATGGTGAAACCCCGTCTCTACTAAAAATACAAAAATTAGCCGGGCGTGGTGGCGGGCGCCTGTAATCCCAGCTACTCGGGAGGCTGAGGCAGGAGAATCGCTTGAACCCGGGAGGCGGAGGTTGCAGTGAGCCGAGATCGCGCCACTGCACTCCAGCCTGGGCGACAGAGCGAGACTCCGTCTCAAAAAAAAAAAAAAAAAAAAAAAAAAAAAA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| AluSx1 | DOF5.1 | 292 | 310 | + | 15.65 | AAAAAAAAAAAAAAAAAAA |
| AluSx1 | DOF5.1 | 293 | 311 | + | 15.65 | AAAAAAAAAAAAAAAAAAA |
| AluSx1 | DOF5.1 | 294 | 312 | + | 15.65 | AAAAAAAAAAAAAAAAAAA |
| AluSx1 | DOF3.6 | 280 | 300 | - | 15.64 | TTTTTTTTTTTTTTTTTTGAG |
| AluSx1 | luna | 140 | 148 | - | 15.26 | ACCACGCCC |
| AluSx1 | ZNF135 | 206 | 219 | - | 15.26 | CCTCCGCCTCCCGG |
| AluSx1 | Klf3/8/12 | 140 | 148 | - | 15.24 | ACCACGCCC |
| AluSx1 | KLF3 | 140 | 149 | - | 14.83 | CACCACGCCC |
| AluSx1 | ERF057 | 209 | 222 | - | 14.79 | CAACCTCCGCCTCC |
| AluSx1 | Nr1h3::Rxra | 58 | 73 | - | 14.65 | TGACCTCAGGTGATCC |
TFBS enrichment in GRCh38
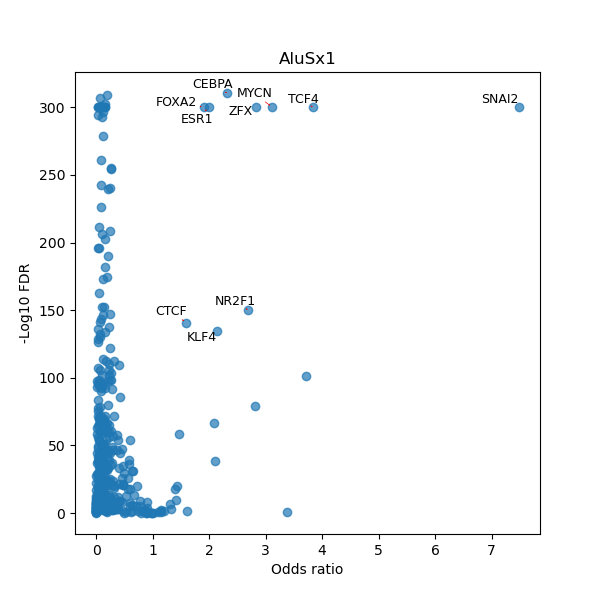
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.