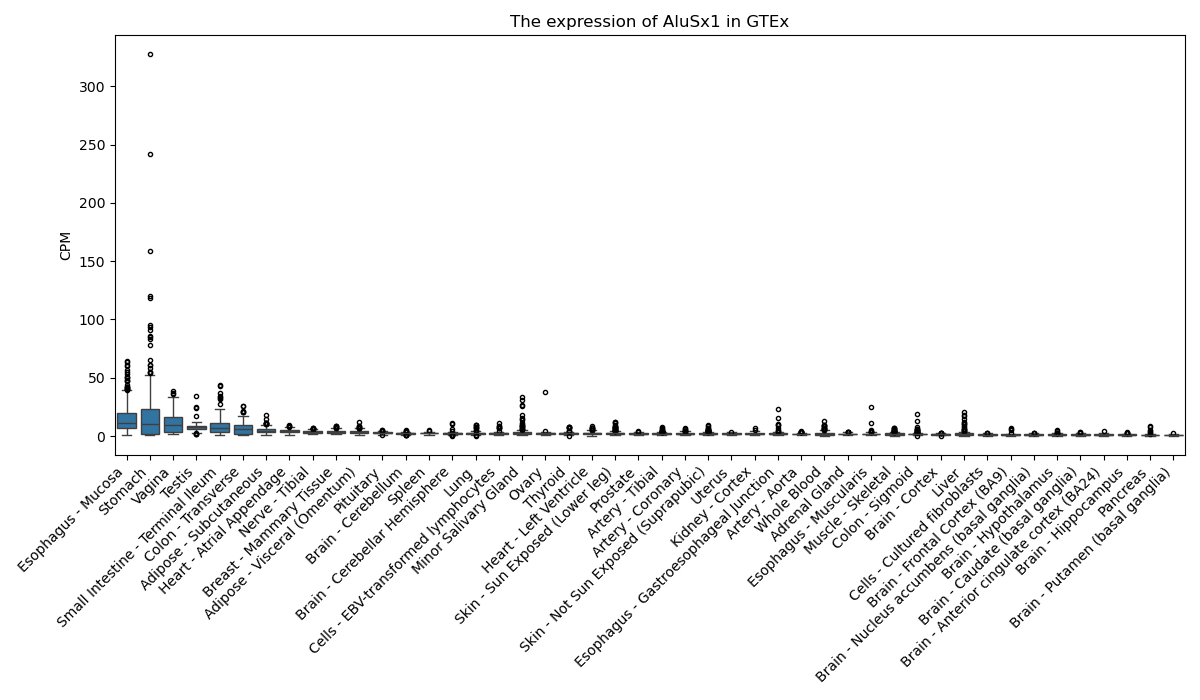
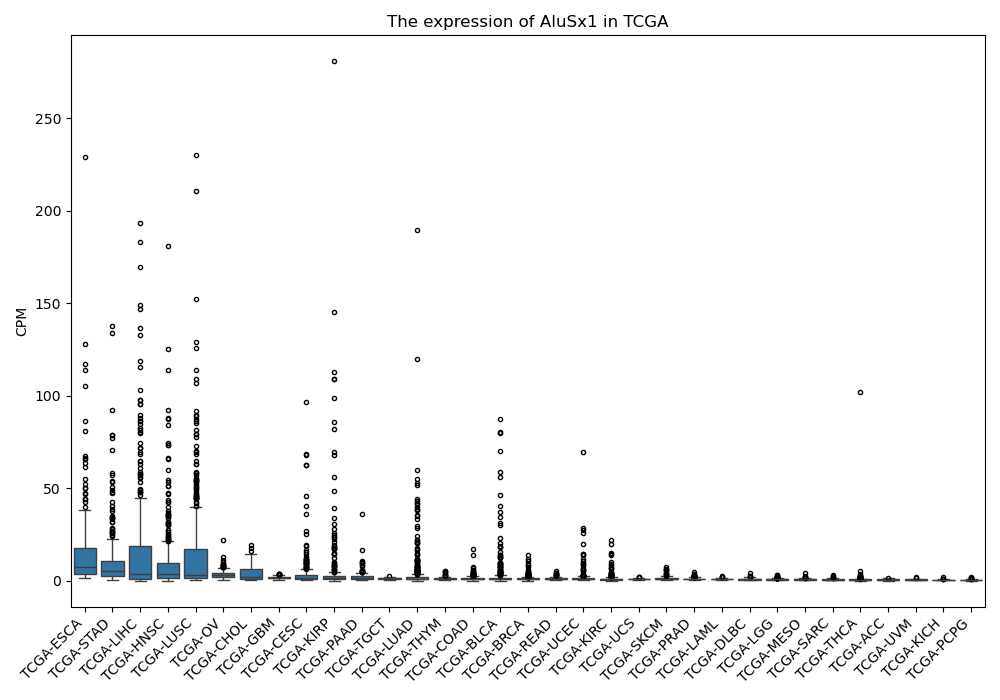
AluSx1
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000048 |
|---|---|
| TE superfamily | Alu |
| TE class | SINE |
| Species | Haplorrhini |
| Length | 312 |
| Kimura value | 7.10 |
| Tau index | 0.7579 |
| Description | AluSx1 subfamily |
| Comment | Alu elements derive from a pair of internally-deleted 7SL RNA monomers that are joined by an A-rich linker. |
| Sequence |
GGCCGGGCGCGGTGGCTCACGCCTGTAATCCCAGCACTTTGGGAGGCCGAGGCGGGCGGATCACCTGAGGTCAGGAGTTCGAGACCAGCCTGGCCAACATGGTGAAACCCCGTCTCTACTAAAAATACAAAAATTAGCCGGGCGTGGTGGCGGGCGCCTGTAATCCCAGCTACTCGGGAGGCTGAGGCAGGAGAATCGCTTGAACCCGGGAGGCGGAGGTTGCAGTGAGCCGAGATCGCGCCACTGCACTCCAGCCTGGGCGACAGAGCGAGACTCCGTCTCAAAAAAAAAAAAAAAAAAAAAAAAAAAAAA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| AluSx1 | DOF3.6 | 290 | 310 | - | 21.52 | TTTTTTTTTTTTTTTTTTTTT |
| AluSx1 | DOF3.6 | 291 | 311 | - | 21.52 | TTTTTTTTTTTTTTTTTTTTT |
| AluSx1 | DOF3.6 | 292 | 312 | - | 21.52 | TTTTTTTTTTTTTTTTTTTTT |
| AluSx1 | DOF3.6 | 282 | 302 | - | 19.33 | TTTTTTTTTTTTTTTTTTTTG |
| AluSx1 | ZNF135 | 39 | 52 | - | 19.08 | CCTCGGCCTCCCAA |
| AluSx1 | ZNF213 | 250 | 261 | - | 18.09 | GCCCAGGCTGGA |
| AluSx1 | DOF3.6 | 281 | 301 | - | 17.41 | TTTTTTTTTTTTTTTTTTTGA |
| AluSx1 | THRA | 57 | 74 | - | 16.59 | CTGACCTCAGGTGATCCG |
| AluSx1 | ZNF135 | 174 | 187 | - | 16.04 | CCTCAGCCTCCCGA |
| AluSx1 | ZNF135 | 71 | 84 | - | 15.84 | TCTCGAACTCCTGA |
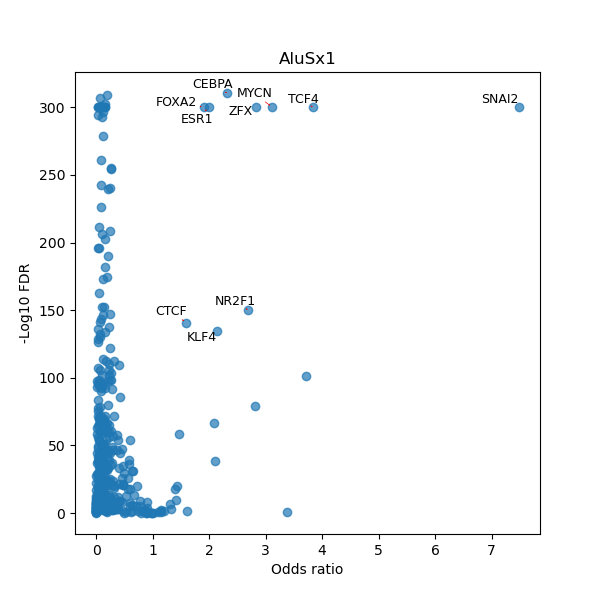
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.